
This is an Application Brief and does not contain a detailed Experimental section.
In this application brief, we present the precise large-scale, label-free analysis of high-density proteomic LC-MS data combined with a qualitative protein strategy that accurately identifies proteins and software that accurately quantifies proteins – both with high reproducibility. The depth to which a complex sample can be interrogated with minimal technical variation is crucial as this characterizes the lowest abundance limit of proteins that can be quantified. This is of equal importance for the software that analyzes LC-MS data since it ultimately defines protein and peptide false quantification rates. ProteinLynx GlobalSERVER and Progenesis LC-MS have been integrated, enabling highly accurate, label-free, relative protein quantification.
Obtain highly accurate, label-free protein quantification with informatics solutions for complex peptide data.
Large scale LC-MS-based discovery experiments that investigate increasingly complex proteomic samples are conducted to assess variation (either experimental or biological), profile samples, or to quantitatively gauge differences in protein abundance. These factors place a demand on the performance of the analytical LC-MS system, as well as on the bioinformatics software required to analyze the data. Robust peak detection, normalization, and alignment of multiple LC-MS runs in larger scale studies are required as this will determine the lowest possible amount of proteins that can be resolved. Results from the SYNAPT G2-S MS for differentially spiked protein standards in a complex cell lysate of Escherichia coli were used to demonstrate the application of Progenesis LC-MS.
Three replicates of two Escherichia coli samples, differentially spiked with bovine serum albumin, alcohol dehydrogenase, enolase, and glycogen phosphorylase B were analyzed. The injected on-column amounts for the spiked proteins in the first sample were 500 attomoles each and 4,000, 500, 1,000, and 250 attomoles for the second sample, respectively. The peptides were separated and analyzed using a Waters nanoACQUITY UPLC System coupled with a SYNAPT G2-S operating at a precursor and product ion mass resolution of > 20,000 FWHM, with data acquired in LC-MSE scanning mode. Searches were conducted with ProteinLynx GlobalSERVER v.2.5.2 and quantified with Progenesis LC-MS.
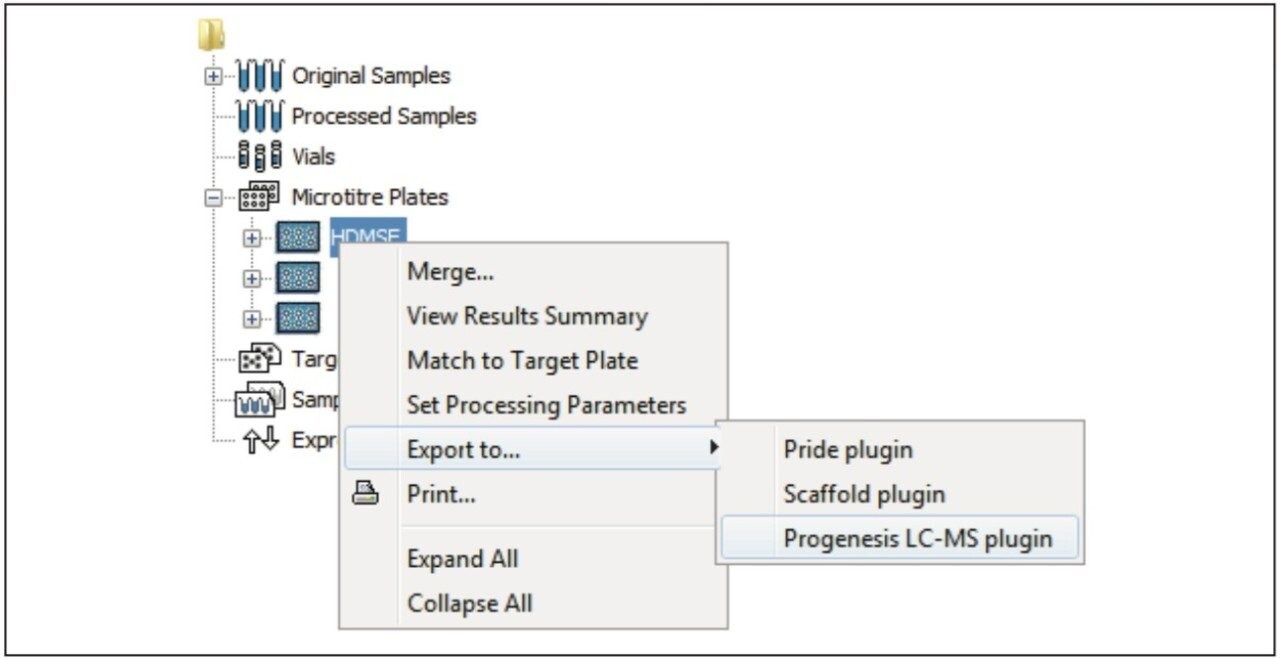
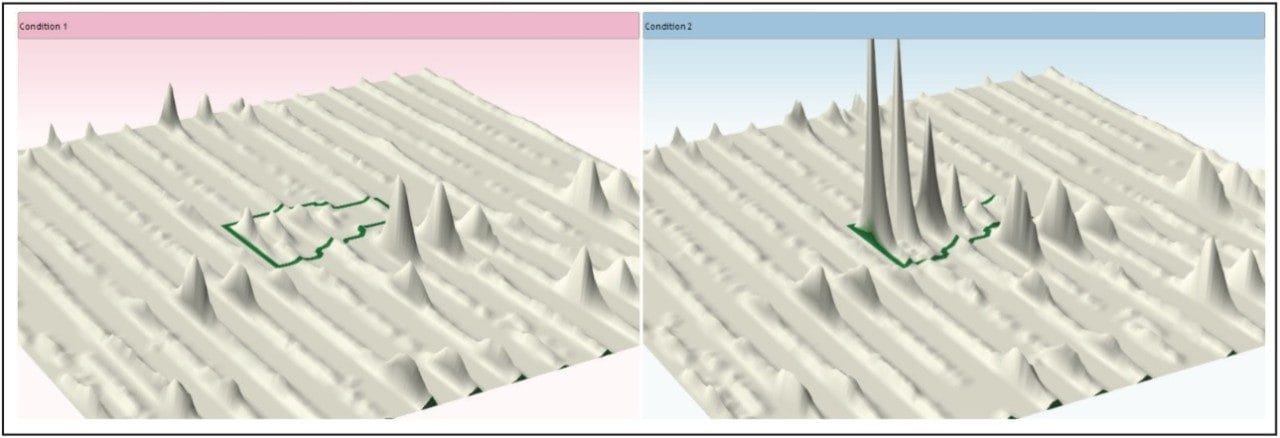
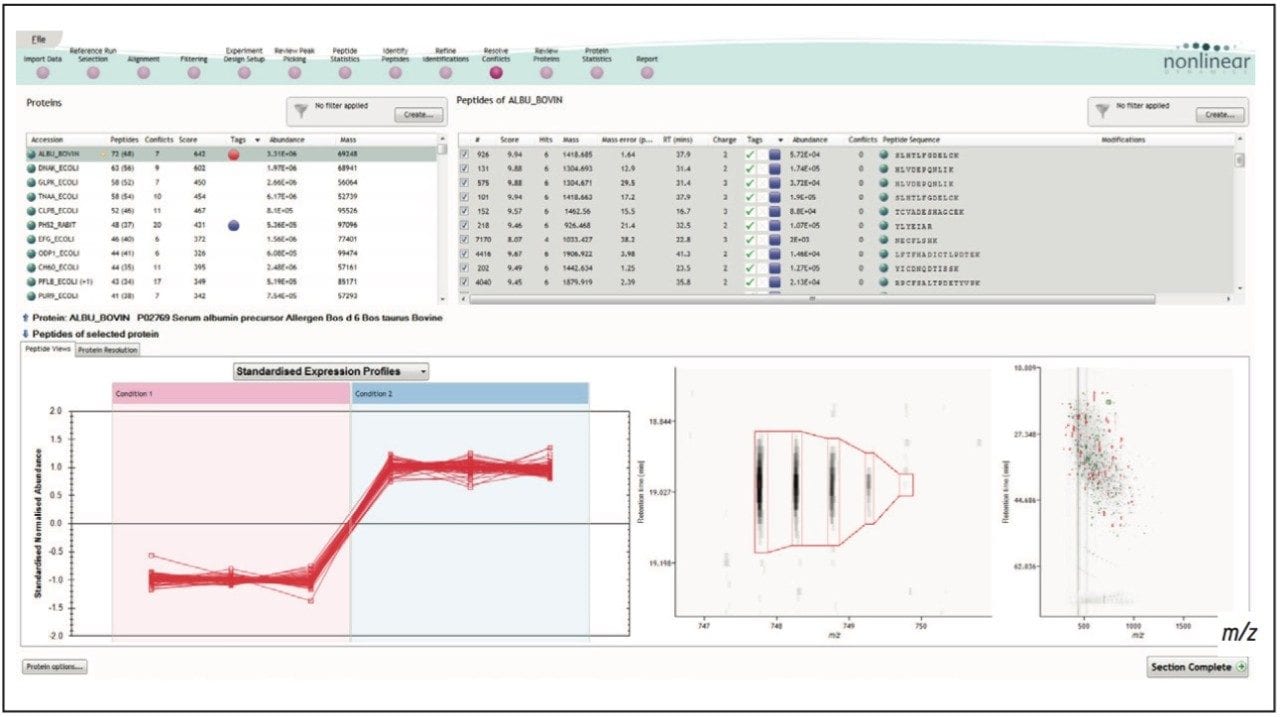
Figure 1 illustrates the plug-in export tool of ProteinLynx GlobalSERVER, which is capable of automatically creating one or more Progenesis LC-MS compatible data files. Visualization of the raw data in 3D-montage view is shown in Figure 2, which confirmed the increased on-column of one of the proteins in the second sample. In this instance, one of the isotopic distributions for one of the charge states of an up-regulated bovine serum albumin peptide is shown. A protein centric summary is shown in Figure 3, which details the peptides of the selected protein, a normalized regulation distribution for the six samples analyzed, a 2D-montage view of the aggregate run, and a fullrange contour LC-MS image of the latter highlighting the peptides of interest.
The label-free relative quantification of four protein standards spiked into a biological background has been demonstrated, using a SYNAPT G2-S operating in LC-MSE mode of acquisition and the results quantified and visualized using Progenesis LC-MS.
720004218, February 2012