This is an Application Brief and does not contain a detailed Experimental section.
The identification and quantification of peptides and proteins from complex proteomic samples has become increasingly important in order to understand function. In this application brief we analyze a Saccharomyces cerevisiae (yeast) sample using a label-free quantitative approach that implements the Hi3 strategy on the nanoACQUITY UPLC System coupled with Xevo G2 QTof. This approach provides increased coverage and confidence in the results due to improved mass resolution and high mass accuracy.
Quantify over 5000 peptides with a dynamic range of three orders of magnitude in a complex mixture using LC-MSE on the Xevo G2 QTof.
The aim of most bottom-up proteomics experiments is to identify and increasingly quantify as many proteins as possible over a wide dynamic range of concentration. LC-MSE on the Xevo G2 QTof is ideal for this type of analysis, as this time-of-flight mass spectrometer provides wide in-spectrum dynamic range, as well as high sensitivity, mass accuracy and resolution, along with confidence in the correct protein identification.
Proteins extracted from a cell lysate of a Saccharomyces cerevisiae (yeast), were digested with trypsin, to produce a complex mixture of peptides. These were separated using a nanoACQUITY UPLC System with a linear gradient from 1% to 40% acetonitrile (+0.1% formic acid) with the eluting peptides analyzed by LC-MSE to identify and quantify the proteins.1
Waters Xevo G2 QTof coupled with nanoACQUITY UPLC, and ProteinLynx Global SERVER v. 2.4 Software were used to analyze triplicate injections of a 400 ng load of yeast cell lysate digest. A typical UPLC gradient from 1.0% to 40.0% over 90 min was used to separate peptides. The mobile phases used were 0.1% formic acid in water and 0.1% formic acid in acetonitrile.
The sample was spiked with 50-fmoles rabbit glycogen phosphorylase B (P00489) per injection to determine the absolute amounts for each of the identified proteins using the Hi 3 method published previously.2
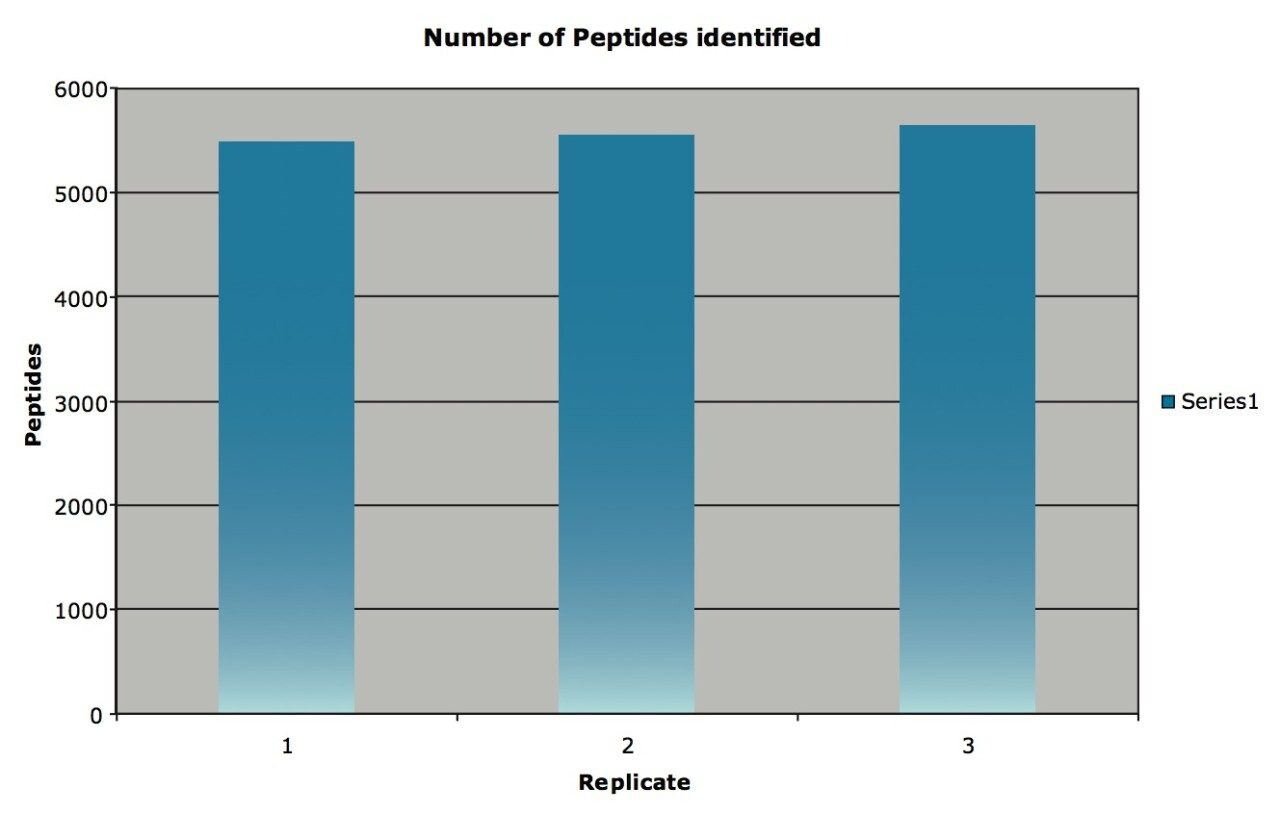
From the analysis of the yeast lysate a total of 6641 unique peptides were identified. 5486, 5553, and 5647 peptides were identified in each replicate respectively, as shown in Figure 1. The mean mass measurement accuracy (of all the pass one peptides) was 1.25, 1.21 and 1.24 ppm respectively for the three replicates.
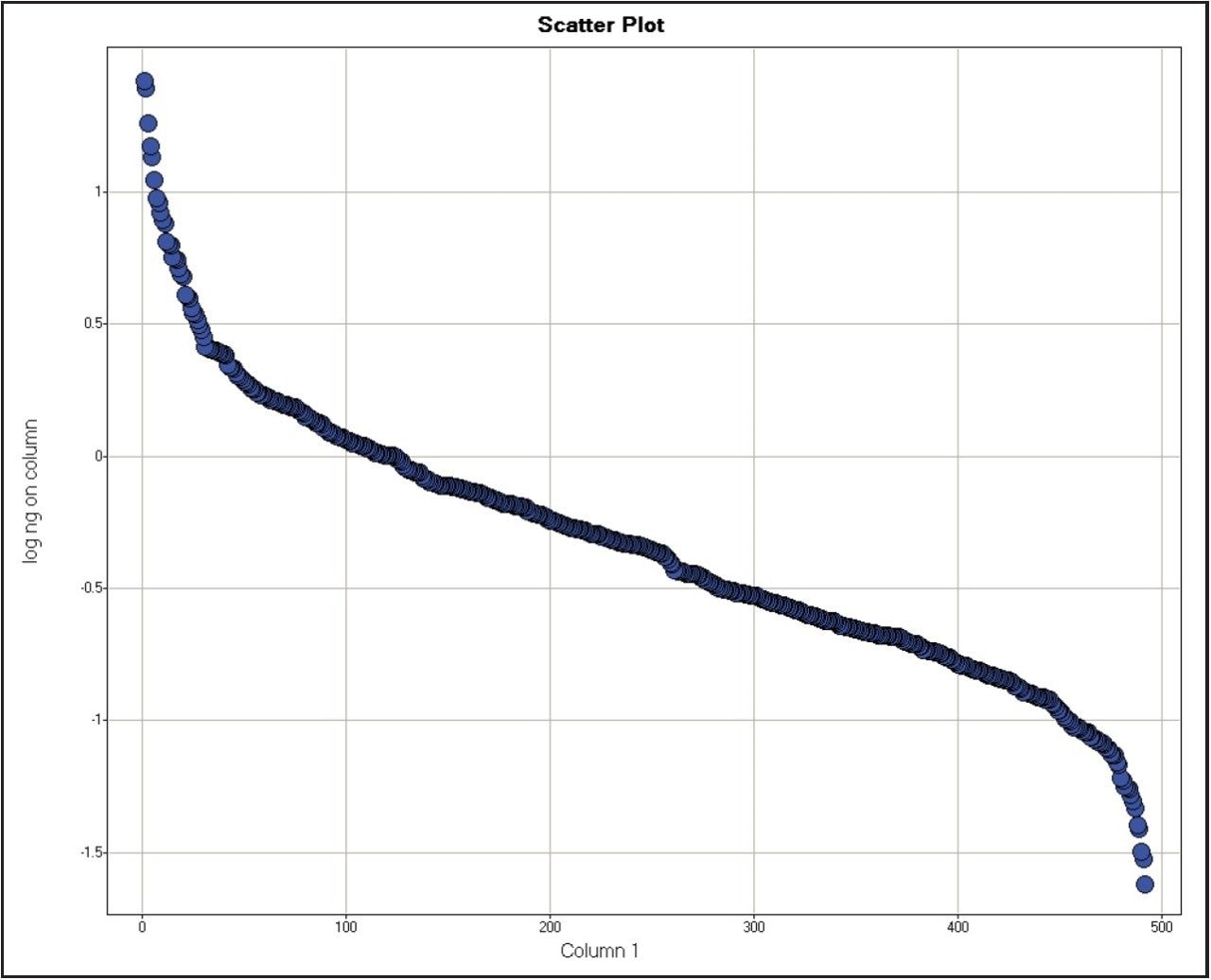
The excellent mass accuracy allowed quantification of 465, 474, and 438 proteins respectively, with 11.8, 11.7, and 12.9 peptides on average matched per protein. Figure 2 shows the dynamic range of concentration measured in the sample from 0.024 to 26.450 ng on column (0.0701 to 526.8200 fmoles on column).
Analysis of a yeast digest sample using MSE LC-MS/MS nanoACQUITY UPLC separation and detection by Xevo G2 QTof, was successful in quantifying over 5000 peptides with a dynamic range of three orders of magnitude in a complex mixture.
720003473, May 2010