This is an Application Brief and does not contain a detailed Experimental section.
This application brief demonstrates to successfully identify a non-targeted unknown contaminant discovered in natural river water during broad scope pesticide screening of environmental waters using TOF/MS.
The precise and reproducible exact mass data acquired by the Xevo G2 QTof offers analysts a non-targeted contaminant screening and discovery solution, with a high degree of confidence in the final result.
TOF screening is frequently used for targeted screening activities where an extensive database is used to target key compounds of interest during the screening acquisition. When analyzing environmental waters, pesticide contamination screening is one of the most important analyses carried out. However, other contaminant species, such as veterinary drugs or human pharmaceuticals and their metabolites may also be present at similar ultra-trace levels as pesticides and could be equally as harmful to the aquatic ecosystem.
Discovery of a non-targeted compound subsequently entails the confirmation and identification of that compound. The TOF instrumentation must be sufficiently sensitive and accurate to ensure that the unknown compound is correctly detected and identified, while at the same time maintaining exact mass accuracy for components at very low concentrations. Accurate and precise exact mass data on both the low energy precursor ion and the MSE high energy fragment ions, along with narrow chromatographic extraction windows, provide increased confidence in the identification of the non-targeted species.
Waters Xevo G2 QTof, coupled with ACQUITY UPLC and ChromaLynx XS Software data processing, were used to rapidly screen natural river water that had been extracted using Oasis HLB Cartridges. A generic screening UPLC gradient was used, with a total runtime of five minutes. The mobile phases used were 10 mM ammonium acetate solution in water and 10 mM ammonium acetate in methanol.
River water blank matrix was screened to investigate any background contamination that might be present. Following deconvolution by ChromaLynx XS Software, a significant peak was discovered at 2.44 minutes, for the ion m/z 237.1031, as illustrated in Figure 1.
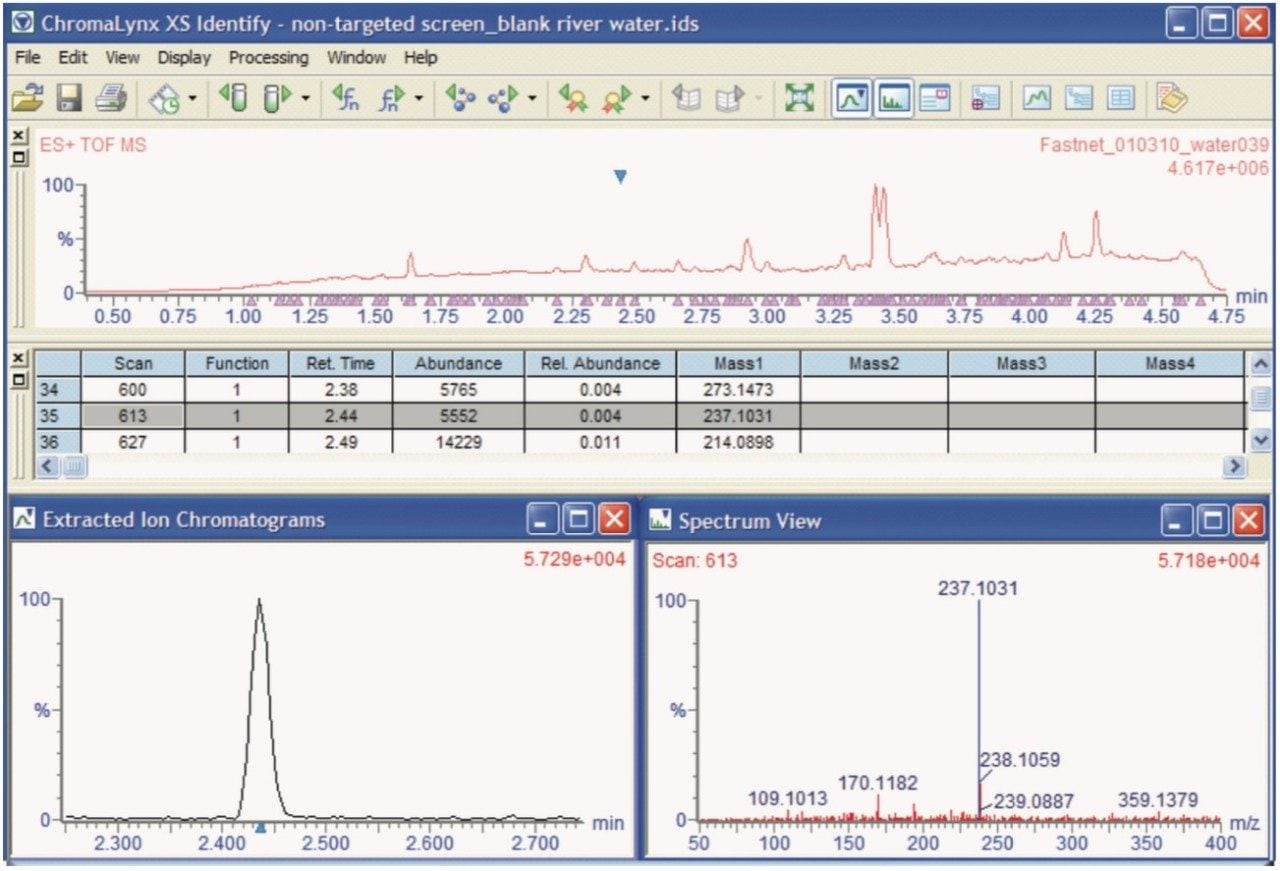
When this exact mass ion was analyzed using the Elemental Composition tool within the MassLynx Application Manager, the formula C15H13N2O was proposed as the most likely, with a maximum mass tolerance of 2.0 ppm, and selected as the best fit using i-FIT. This formula matched that of protonated carbamazepine, a human anti-convulsant and mood stabilizing drug. Both the low energy MS and the high energy MSE spectra acquired at 2.44 minutes were then processed using the MassFragment tool, and matched to the parent molecule of carbamazepine and its primary fragment ion, as shown in Figure 2.
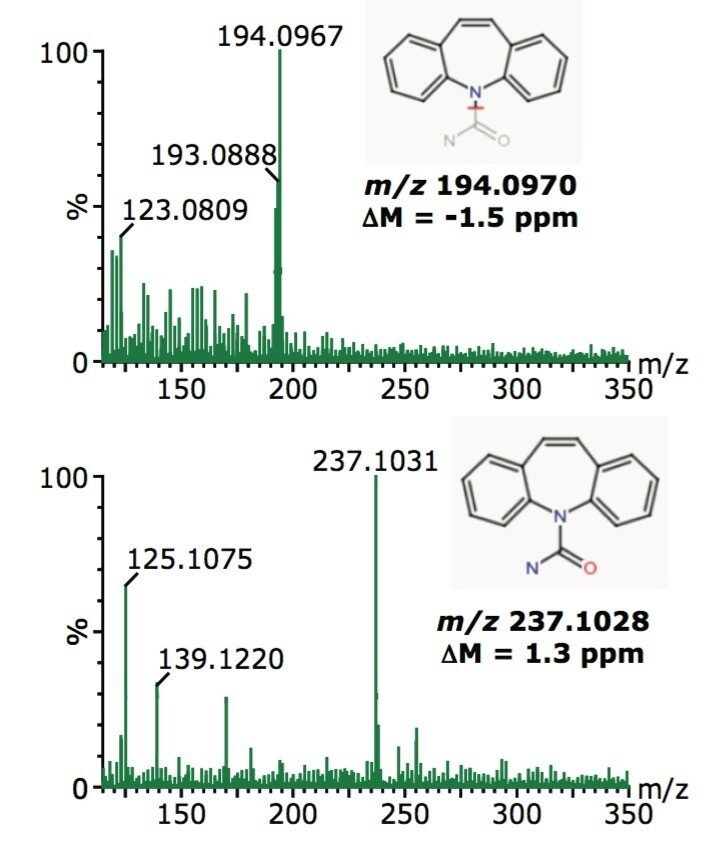
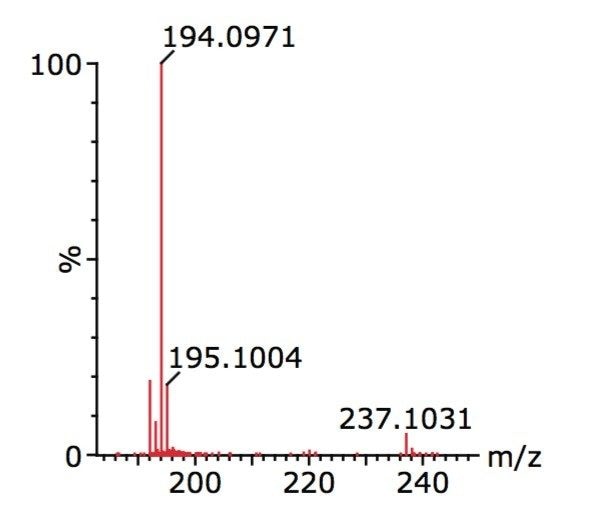
Finally, unequivocal confirmation was made by comparison with a solvent standard solution of carbamazepine alone. The solvent standard data, shown in Figure 3, established a match to the nontargeted contaminant data, clearly demonstrating that the unexpected compound is carabamazepine.
Oasis HLB SPE extraction with rapid ACQUITY UPLC separation and detection by Xevo G2 QTof, followed by data processing using ChromaLynx MS Software, was successfully used to screen natural river water.
A non-targeted screening approach was used and enabled the detection and identification of an unexpected contaminant, the drug molecule carbamazepine.
The precise and reproducible exact mass data acquired by the Xevo G2 QTof allowed unequivocal assignment of structures to both the precursor and fragment ions. This approach offers analysts a non-targeted screening and discovery solution, with a high degree of confidence in the final result.
720003424, April 2010