For research use only. Not for use in diagnostic procedures.
The heavily glycosylated SARS-CoV-2 spike protein is implicated in COVID-19 infectivity and consequently numerous efforts to characterize the glycosylation patterns of the capsid spike protein are being pursued in an effort to guide vaccine and immunotherapy development. Here, the unique configuration of the novel Waters SELECT SERIES Cyclic IMS instrument is exploited for characterization of the glycan structures and linkages of O-linked glycopeptides found near the furin protease cleavage site of the spike protein; furin cleavage exposes the fusion sequence of the spike protein that results in separation of the S1 and S2 subunits is thought to be needed for cell entry. Additionally, extended core 1 and core 2 type structures with both α2-3 and α2-6 sialic acid linkages are identified; glycosylation has been shown to alter substrate recognition and the ratios of the core 2 and extended core 1 structures may impact the virulence of the novel coronavirus.
The unique design of the Cyclic IMS system enables site-specific, linkage-specific characterization of the glycan structures of low-level O-linked glycopeptides from the SARS-CoV-2 spike protein. Trap fragmentation is used to release oxonium ion fragments from the O-glycan moiety and high-resolution ion mobility is used to separate and analyze the fragment ions.
The outbreak of the recent COVID-19 pandemic, caused by the SARS-CoV-2 coronavirus, has resulted in the sickening of millions and the deaths, to date, of over 2 million people globally.1 Because of its relative novelty, the scientific community has been engaged in rapid characterization and compilation of the biophysical characteristics of the virus, in hopes of aiding the development of effective vaccine and immunotherapies. The SARS-CoV-2 virion enters the cell by binding to the host cell ACE2 (angiotensin-converting enzyme 2) receptor with a protrusive transmembrane spike protein.2 The SARS-CoV-2 coronavirus spike protein is a homotrimeric class I fusion protein consisting of two heavily glycosylated subunits, S1 and S2.3,4 Previous studies of known viral pathogens, including influenza, have revealed the glycan composition of the outer envelope plays a key role in immunoevasion, notably through steric hindrance of immunorecognition sites.5–7
The characterization effort of the SARS-CoV-2 pathogen has included several comprehensive studies of the glycosylation of the coronavirus spike protein. To date, these studies have indicated consensus occupancy of 14 of the 22 available N-linked glycosylation sites and conflicting occupancy of the remaining seven sites.8–16 In addition, Shajahan et. al showed evidence for three O-linked glycosylation sites on the S1 domain, one of which was confirmed by Sanda et. al.10,11 In a sequence analysis of the SARS-CoV-spike protein, Andersen and co-workers predicted a furin cleavage site, a site critical to protein activation, in the linker region flanking the S1 and S2 subunits.2 This region containing the furin cleavage site was further predicted to contain up to three additional O-linked glycosylation sites, the function of which was speculated to be related to infectivity and transmissibility.2 Characterization of O-linked glycans is notoriously difficult for several reasons: the lack of a consensus sequence, high heterogeneity among the glycoforms, and low relative abundance. Consequently, characterization of these types of analytes requires high performance chromatography and high resolution, high sensitivity mass spectrometry. Previous studies using the Waters SYNAPT platform have shown the capacity to resolve branched and linear glycan structures of isomeric glycans and separate α2-3 and α2-6 linkage isomers from glycan standards.12–16 MS/MS fragmentation with the Waters SELECT SERIES Cyclic IMS provides clear evidence of O-linked glycosylation along the linker region preceding the polybasic furin cleavage site and cyclic ion mobility is used to site-specifically separate and characterize the O-linked glycan structures. The scalable resolution afforded by this unique experiment provides evidence for a mixture of core 1, extended core 1, and core 2 structures, with resolution of NeuAcα2-3Galβ1-3GalNAc and NeuAcα2-6Galβ1-4GlcNAc, isomers that feature highly similar collisional cross sections.
Recombinant SARS-CoV-2 spike protein was expressed in HEK 293 cells and obtained from Acrobiosystems. The disulfide linkages in the spike protein were reduced with DTT and treated with iodoacetamide to alkylate cysteine residues prior to digestion with PNGaseF and trypsin.
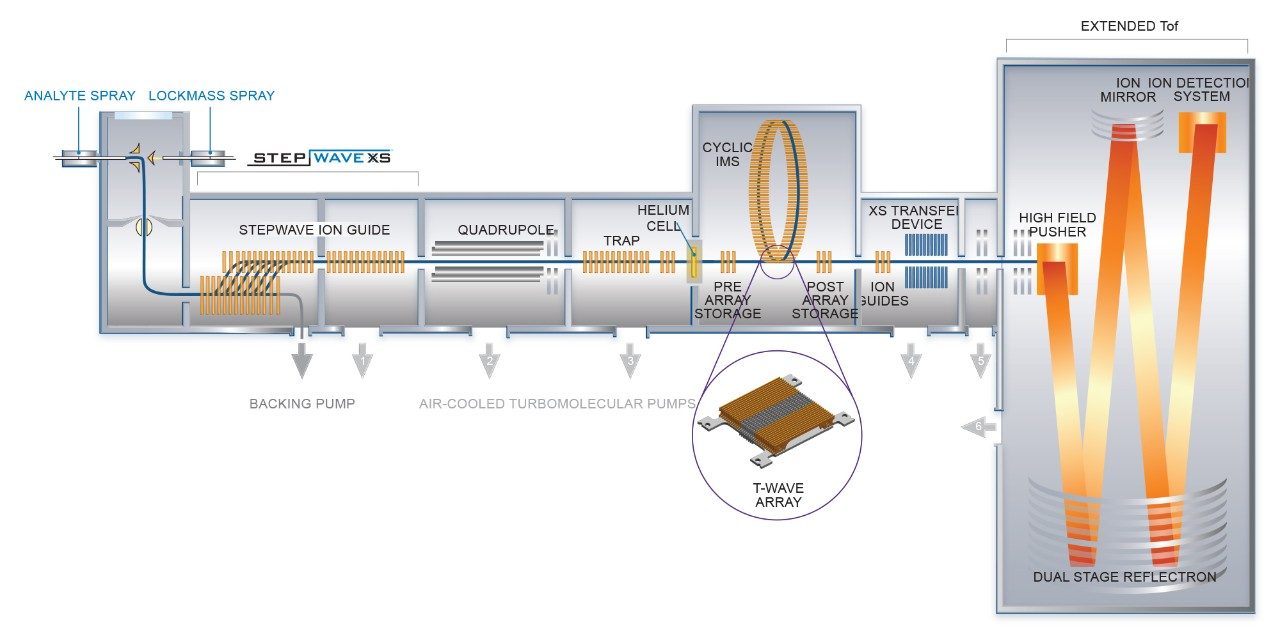
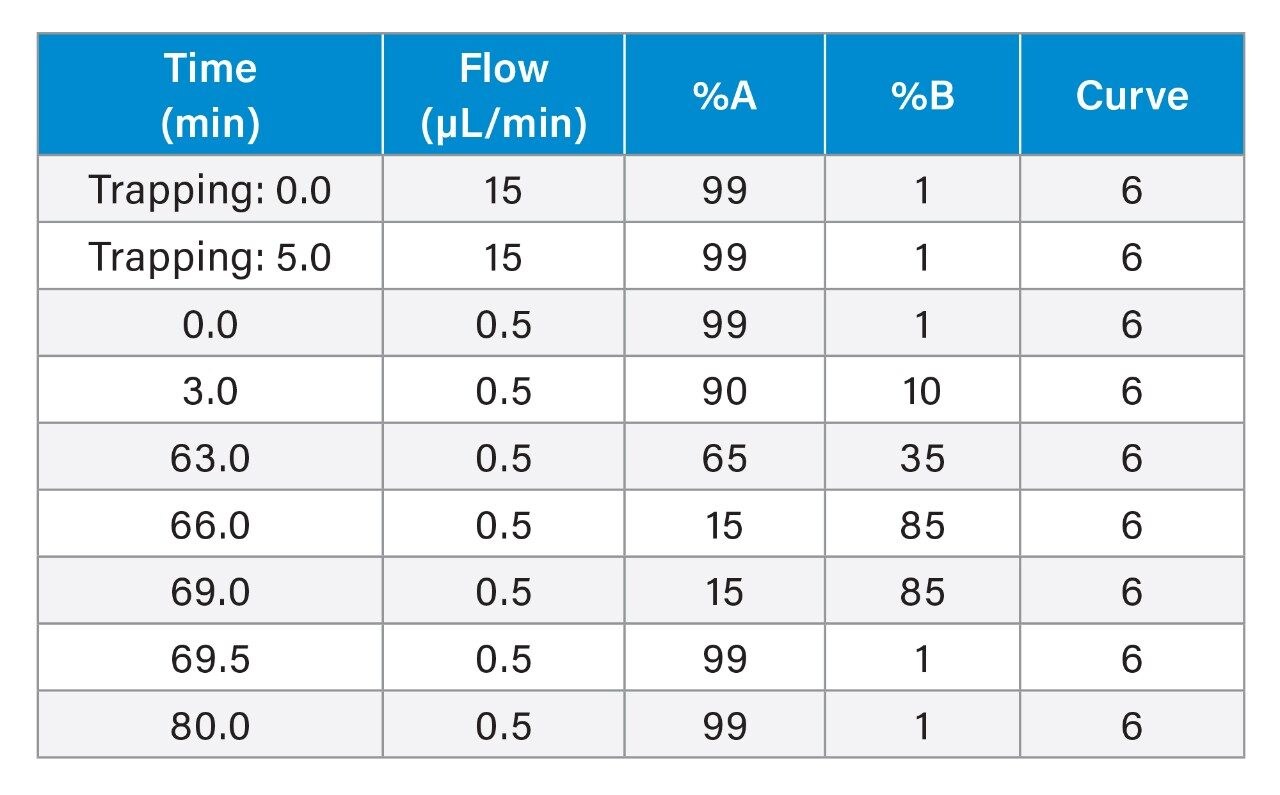
General conditions for the chromatography and mass spectrometry are listed in Tables 1, 2, and 3. Cyclic IMS methods using five passes of the cyclic device were optimized previously for tri- and disaccharide fragments using O-linked glycopeptides from hemopexin. O-glycopeptides from the SARS-CoV-2 spike protein were quadrupole isolated) prior to collisional activation in the trap region of the Cyclic IMS system (shown in Figure 1). Fragments of the glycopeptides were separated using the cyclic ion mobility cell. Methods were optimized such that trisaccharide fragments passed through the device five times before ejection and detection.
|
LC system: |
ACQUITY UPLC M-Class |
|
Detection: |
SELECT SERIES Cyclic IMS Mass Spectrometer |
|
Vials: |
QuanRecovery |
|
Column(s): |
nanoEase M/Z HSS T3 100 Å, 1.8 µm (75 µm x 15 cm) p/n: 186008816 nanoEase M/Z Symmetry C18 100 Å, 5 µm (180 µm x 20 mm) trap, p/n: 186008821 |
|
Column temp.: |
60 °C |
|
Sample temp.: |
8 °C |
|
Injection volume: |
1–5 μL |
|
Flow rate: |
500 nL/min |
|
Mobile phase A: |
H2O, 0.1% FA, 1 ppm citric acid |
|
Mobile phase B: |
ACN, 0.1% FA, 1 ppm citric acid |
|
MS system: |
Cyclic IMS |
|
Ionization mode: |
ESI+ |
|
Acquisition range: |
m/z 50–2000 |
|
Capillary voltage: |
2.8 kV |
|
Collision energy: |
30 V–70 V |
|
Cone voltage: |
20 V |
|
Number of passes: |
5 |
|
Racetrack TW height: |
22 V |
|
Racetrack TW velocity: |
375 m/s |
|
IMS sequence: |
Inject, Separate, Eject, and Acquire |
|
IMS cycle time: |
100 ms |
|
Chromatography software: |
MassLynx SCN1016 Release 3 |
|
MS software: |
Waters Embedded Analyzer Platform for Cyclic IMS Release 4 |
|
Informatics: |
DriftScope 2.9, modified for Cyclic IMS |
Targeted ion mobility MS/MS experiments were performed on the observed O-linked glycoforms of tryptic peptide AGC(carbamidomethyl)LIGAEHVDNSYEC(carbamidomethyl)DIPIGAGIC(carbamidomethyl)ASYQTQTNSPR, denoted T56, using fragmentation in the trap region to generate peptidic and oxonium ion fragments prior to the ion mobility cell. The scaleable resolution of the cyclic ion mobility (cIM) instrument was leveraged to separate isomeric glycan structures following release by collisional activation. Two methods were developed previously for 1 and 5 passes of the cyclic device for trisaccharide fragments and were used to separate isomeric forms of the HexNAc-Hex and HexNAc-Hex-NeuAc oxonium ion fragments of T56-HexNAc(1)-Hex(1)-NeuAc(1), T56-HexNAc(1)-Hex(1)-NeuAc(2), T56-HexNAc(2)-Hex(1)-NeuAc(1), T56-HexNAc(2)-Hex(2)-NeuAc(1), and T56-HexNAc(2)-Hex(2)-NeuAc(2). These peptides are listed in Table 1 as glycopeptides A-D.

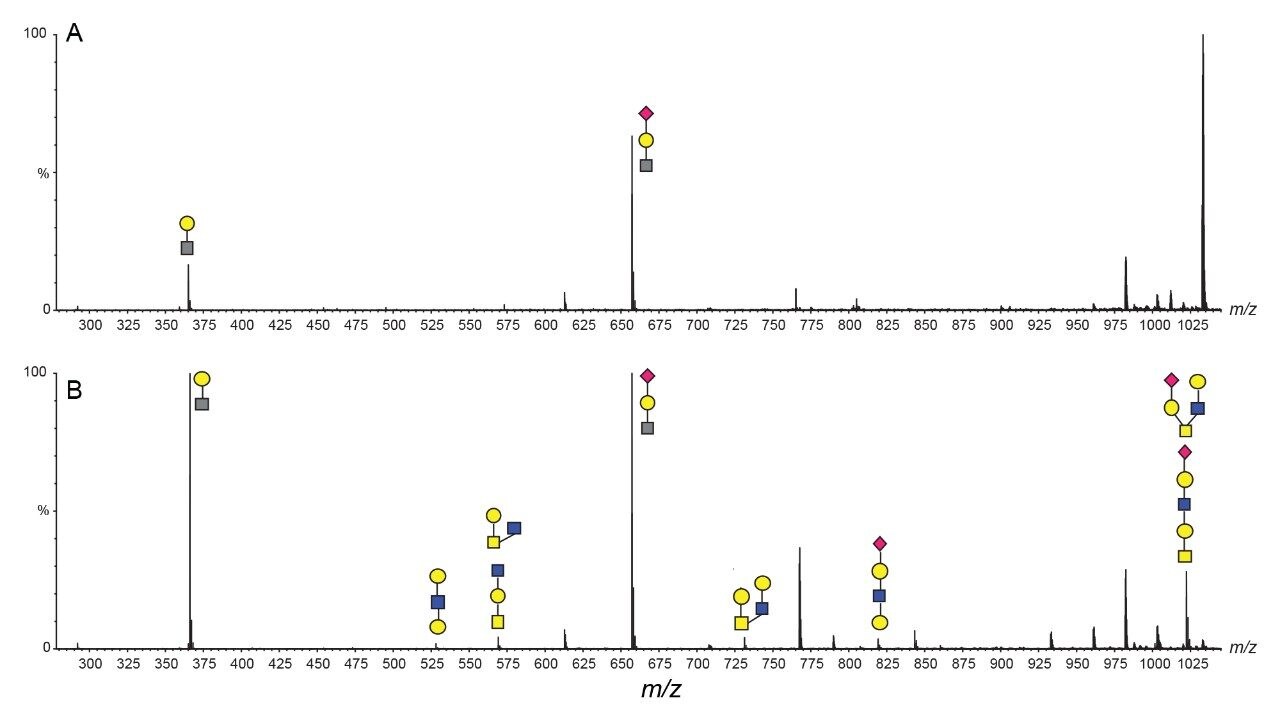
Chromatographic peaks yielding a triply charged unmodified T56 peptide as a fragment were integrated in DriftScope 2.9 and the mobility information was extracted. In general, 2–4 chromatographic peaks were observed for each glycoform, indicating that some isomeric heterogeneity can be separated by reverse phase chromatography. Collision induced fragmentation of the T56 glycopeptides generally yielded a mixture of peptidic, oxonium, and peptide + glycan fragments. Figure 2 shows MS/MS spectra for A) glycopeptide A and B) glycopeptide C, demonstrating a high yield of large oxonium ion fragments, particularly for glycopeptide C. Observation of the HexNAc(2)-Hex(2)-NeuAc(1) oxonium ion fragment at m/z 1022 confirms the presence of a single pentasaccharide glycoform rather than separate tri- and disaccharide components localized at two different sites. Oxonium ions at m/z 528 and 819 confirm the presence of extended core 1 structures.
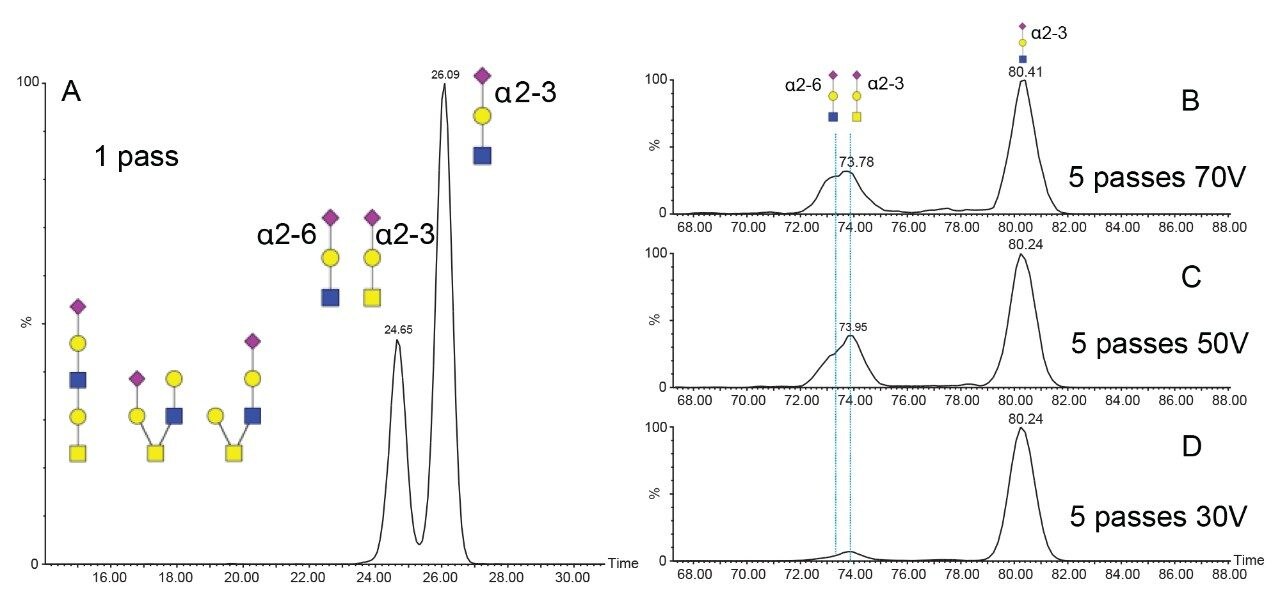
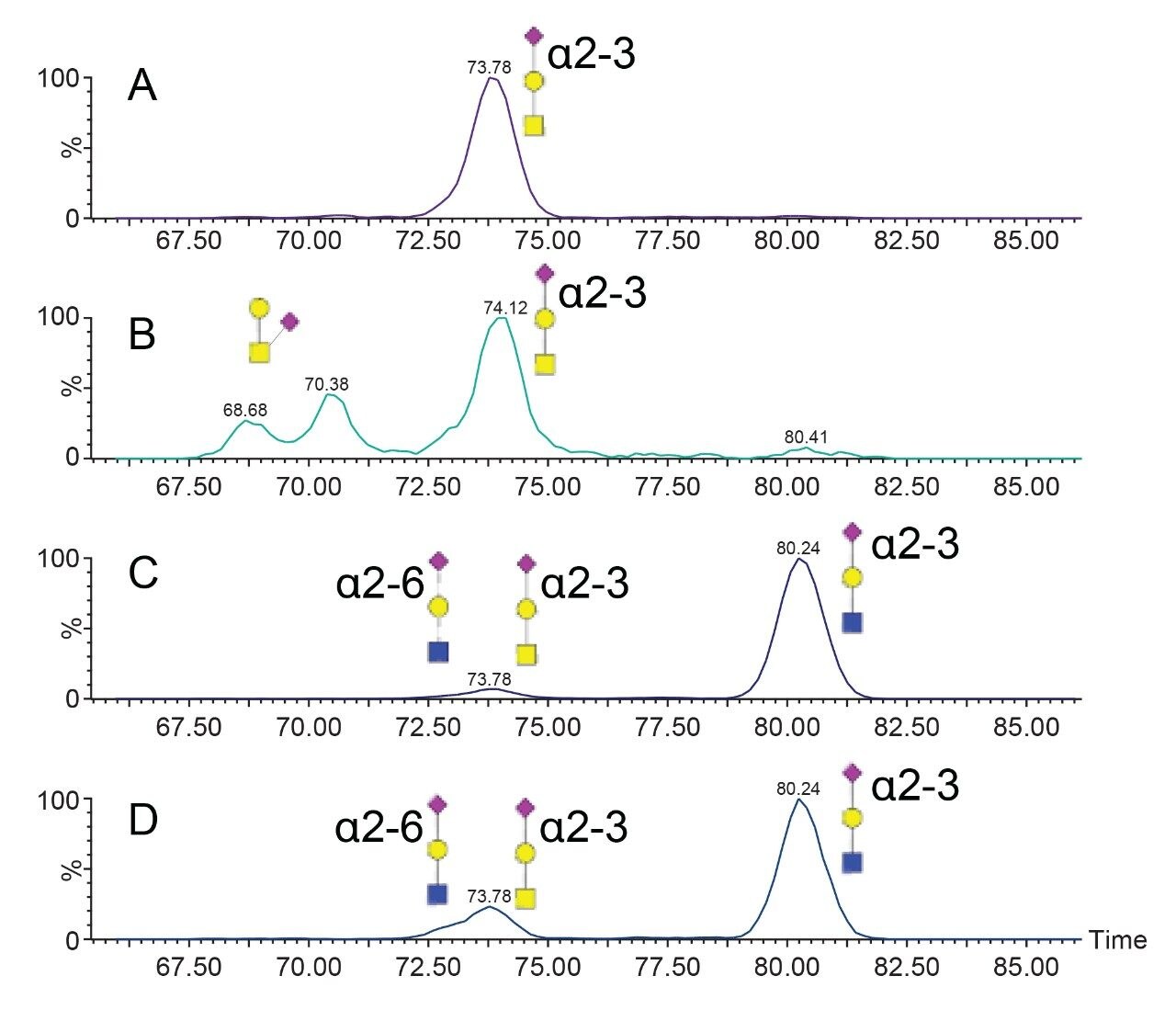
Single pass and multi pass (5 cycles) ion mobility experiments were undertaken for the HexNAc-Hex-NeuAc oxonium ion. The resulting arrival time distributions are shown in Figure (3B-D) for glycopeptide C using variable collision energies. Using a single cyclic IM pass, two mobility populations are evident; using five passes, the more compact species further separates into two partially resolved isomers. Collisional cross sections were calculated for the 1 pass using the Major Mix IMS calibration standard as a reference. The two arrival time distributions observed with 1 pass have calculated collisional cross sections of 234.1 and 245.4 Å2, respectively. Guttman et. al. previously measured CCS values for NeuAcα2-6Galβ1-4GlcNAc and NeuAcα2-3Galβ1-4GlcNAc as 236.9 Å2 and 247.2 Å2, respectively.12 The most elongated measured HexNAc-Hex-NeuAc isomer is in good agreement with a NeuAcα2-3Galβ1-4GlcNAc. Using 5 passes of the cyclic mobility device, the single mobility peak observed at 234.1 Å2 was resolved into two substructures. Previous studies have shown alpha 2–6 sialic acid linkages to be more stable than alpha 2–3 linkages; thus, experiments in which the collision energy was varied are shown in Figure 3 (B–D).17 Higher collision energy yields an increase in the relative abundance of the most compact of the three isomers resolvable with 5 passes. Core 1 type structures (NeuAcα2-3Galβ1-3GalNAc) are the most commonly observed for O-linked glycopeptides; thus, the more elongated and less stable of the compact isomers in the 5-pass separation is consistent with a NeuAcα2-3Galβ1-3GalNAc, type 1 core structure. The more compact and less stable is assigned to a NeuAcα2-6Galβ1-4GlcNAc, which would be expected from fragmentation of a core 2 structure.
Five pass cyclic ion mobility was applied to the m/z 657 (HexNAc-Hex-NeuAc) oxonium ion from the remaining glycopeptides and is shown in Figure 4. The glycopeptides having two HexNAc residues (C and D) yielded similar ion mobility arrival time distributions (ATDs), suggesting a mixture of extended core 1 and core 2 structures, as shown in Figure 3. The simplest of the glycopeptides, having a single HexNAc, Hex, and NeuAc residue (A), formed a single mobility distribution at 73.8 ms, which was assigned to NeuAcα2-3Galβ1-3GalNAc for the T56-HexNAc(2)-Hex(2)-NeuAc(1) glycopeptide and is consistent with known glycobiology. In contrast, separation of the m/z 657 oxonium ion from the T56-HexNAc(1)-Hex(1)-NeuAc(2) glycopeptide resulted in two additional mobility peaks at 68.7 and 70.4 ms. These are likely branched structures in which the NeuAc residue is linked to the GalNAc by either 2–3 or 2–6 linkages. These peaks were not observed for glycopeptide D, suggesting sialylation of the core GalNAc residue (Figure 4B) is rare for the T56 glycopeptide and occurs only on the simple core 1 structure.
The SELECT SERIES Cyclic IMS is demonstrated to show separation of the oxonium ion fragments of SARS-CoV-2 spike protein O-glycopeptides, providing site-specific and linkage specific information about the glycopeptide sugar structures. The high sensitivity, scalable ion mobility resolution, and unique geometry of the Cyclic IMS instrument allows for targeted CID-cIM-MS experiments of low-level O-glycopeptides, affording site-specific structural characterization of highly heterogeneous proteins. This strategy provides improved detail for the ongoing, global effort to characterize, understand, and ultimately target the SARS-CoV-2 virus.
720007081, Revised January 2021