
This is an Application Brief and does not contain a detailed Experimental section.
This application brief demonstrates the applicability of the ACQUITY UPLC with the Xevo TQ-S for the quantification of peptides derived from allergenic proteins in complex food matrices.
The prevalence of food allergies is continuing to rise, with an increasing amount of the population having an adverse reaction to food. While many foods may provoke reactions, over 90% of food-allergic reactions are limited to a small number of so-called critical allergens. These critical allergens are contained in cow’s milk, crustaceans, eggs, fish, peanuts, soy, tree nuts, and wheat. In order to help those who suffer from food allergies, there are many different regulations that address this food safety issue across the globe.
For many years, the most popular analytical techniques to analyze the presence of allergenic ingredients in food have been antibody based enzyme-linked immunosorbent assay (ELISA) and real-time polymerase chain reactions (PCR). ELISA relies on the availability of antibodies that target the allergenic protein and that do not elicit a response from other similar proteins. PCR does not target the specific allergenic protein of interest and can therefore give rise to false positive results. Recently, the use of mass spectrometry (MS) for the detection of allergens has generated much interest due to its ability to target the specific protein and to analyze multiple allergenic markers in a single method.
In this application brief, an approach for the detection of peptide markers of allergens in a food matrix is demonstrated using peanut in bread. An important advance in instrument design that enables the simultaneous acquisition of matrix background information during the quantitative MRM analysis is also illustrated. This functionality (known as RADAR acquisition) offers an innovative approach for the quantification of analytes of interest while monitoring background interferences. This enables informative decisions to be made during method development and continuous monitoring of the matrix background to ensure a robust and reliable method.
Previous work using a food-proteomics approach to identify markers associated with the allergic reaction induced by ingestion of raw or roasted peanuts was investigated on Waters nanoACQUITY UPLC and Xevo QTof MS (Application note no. 720003656EN). From that work, peptide sequences observed in both raw and roasted peanuts were identified. In the work presented here, two of those peptides have been used to develop a method for highly sensitive and specific detection using the ACQUITY UPLC System with the Xevo TQ-S Mass Spectrometer.
Bread was selected as the food matrix and peanut protein Ara h1 as the example allergenic protein. Proteins were extracted and then digested using trypsin. The selected marker peptides were incorporated into a single UPLC-MS/MS method. Spiked mixtures as well as the un-spiked bread matrix were analyzed using the Xevo TQ-S operated in RADAR mode.
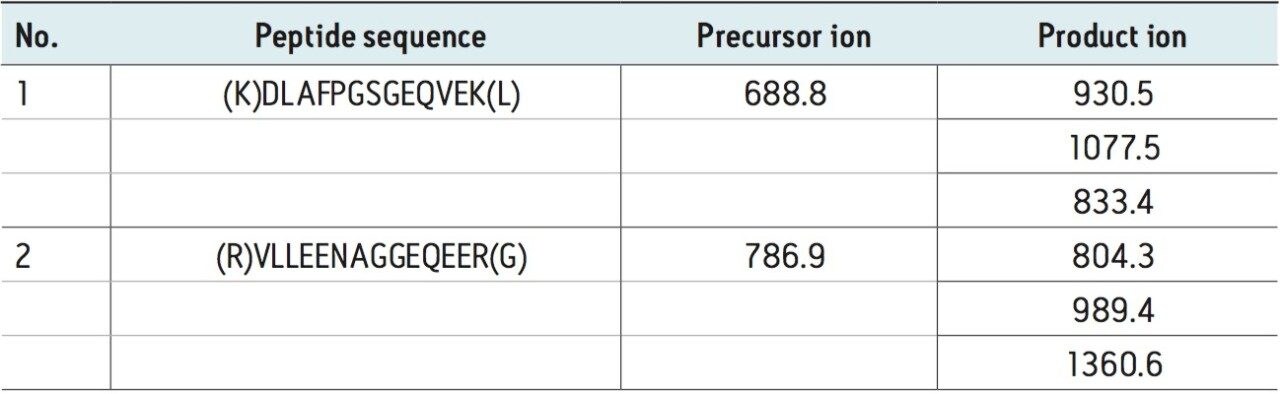
The three MRM transitions selected for each of the candidate peptides of Ara h1 in peanut are listed in Table 1, and the resulting MRM chromatogram for each primary transition is shown in the lower portion of Figure 1. The three extracted MRM transitions for the peptide sequence VLLEENAGGEQEER are shown in the inset.
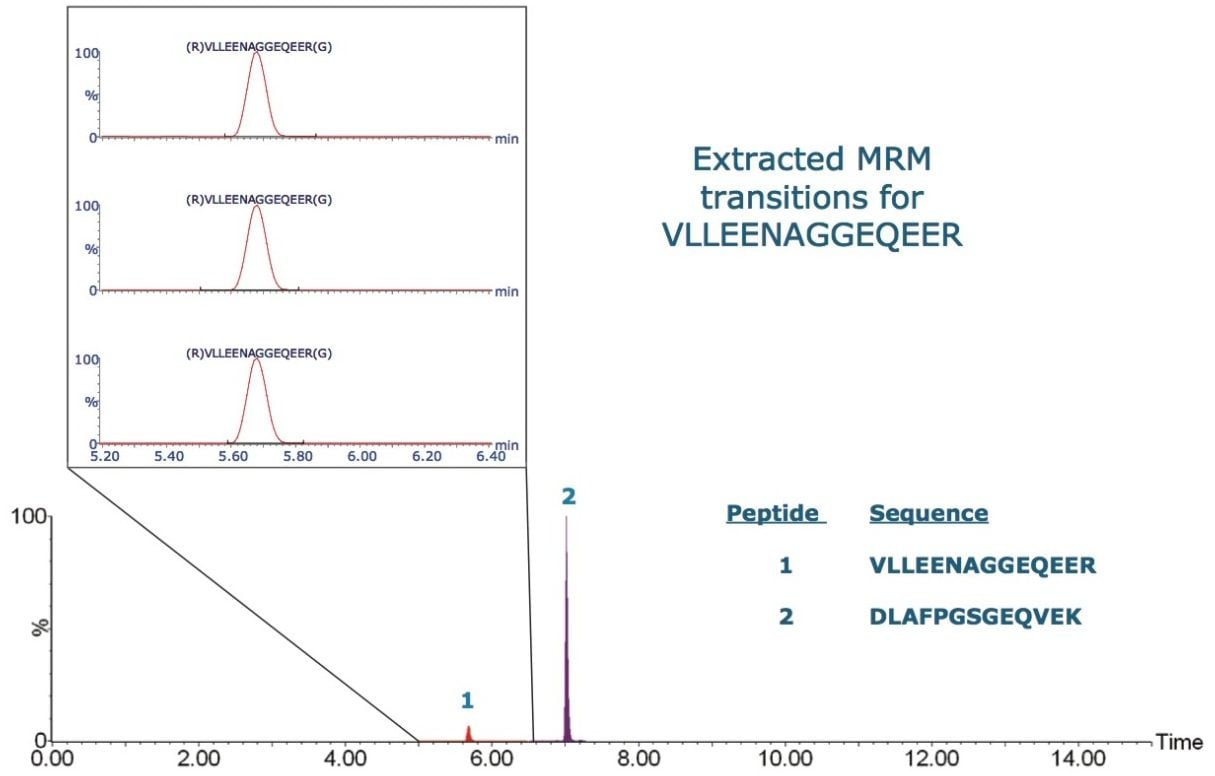
The simultaneous acquisition of full scan data along with the MRM data enabled assessment of the complexity of the matrix throughout the quantitative experiment. Figure 2 shows the full scan total ion current (TIC) chromatogram along with the MRM chromatograms of the individual peptides. For the retention times of each peptide, the summed spectra are shown in the inset.

Figure 2. Background matrix information generated using RADAR.
A: MRM chromatogram of 786.9 > 804.3 (VLLEENAGGEQEER)
B: MRM chromatogram of 688.8 > 930.5 (DLAFPGSGEQVEK)
C: Full scan MS TIC chromatogram.
Insets to A and B show summed full scan spectra at Rt 5.67 and 7.01, respectively.
As can be seen from the RADAR spectra and TIC chromatogram, the digested bread matrix is incredibly complex, with many components eluting at any point throughout the chromatogram. The ion signal from each of the two peptides is highlighted by arrows on the spectra. Even with such a complex matrix, the MRM chromatograms show excellent signal-to-noise for the peptide MRM transitions, demonstrating the excellent sensitivity and selectivity of the Xevo TQ-S.
A rapid and specific UPLC-MS/MS method has been developed to demonstrate the detection of peptide markers of allergens in food matrix using ACQUITY UPLC and Xevo TQ-S. The use of RADAR functionality enabled the simultaneous acquisition of MRMs for quantification and full scan data to monitor the highly complex matrix background. There is great potential to extend this current method into a multi-allergen screening method.
720004103, September 2011