
The purification strategy for single-stranded RNA oligonucleotides presented here is rapid, cost effective, and yields high purity material using the Waters ACQUITY UPLC System with Oligonucleotide Separation Technology (OST) Column chemistry.
Oligonucleotide synthesis is a very efficient and high-yielding process. Typical yields of oligonucleotide reactions carried out on solid support range from 98 to 99.5% per coupling step. In a typical multi-step oligonucleotide synthesis, impurities accumulate and the overall yield of even a modest sized 21-mer oligonucleotide can range from 67 to 90%, with longer chain oligonucleotides giving correspondingly lower yields.
For researchers, it is often necessary to work with materials of higher purity than are available from crude synthetic mixtures. For this reason, oligonucleotides used for gene knockout, genotyping, and diagnostic purposes are typically purified following synthesis. Few economically viable solutions exist for lab-scale purification of oligonucleotides, and those that do exist - such as ion-exchange chromatography and polyacrylamide gel electrophoresis - are often cumbersome and/or time-consuming.
In this application note, we describe a cost-effective and rapid method for the purification of modest quantities of material, up to 140 nmoles in a single injection, with final purities of greater than 95% using the Waters ACQUITY UPLC System with Oligonucleotide Separation Technology (OST) Column chemistry. The purification scale presented matches well with typical oligonucleotide synthetic scales (50 to 250 nmol). The method described below allows for the purification of oligonucleotides with high purity products in 15 to 30 minutes.
The RNA oligonucleotide 5’ - CCU UGU AAU CGC UUG ACG ATT - 3’ was purchased from a vendor and reconstituted in 110 μL of 0.1 M triethylammonium acetate (TEAA) to yield a solution of approximately 2.8 nmol/μL. The sample was prepared immediately prior to use to prevent degradation.
The RNA oligonucleotide was purified using a Waters Alliance HPLC Bioseparations System using a Waters XBridge BEH OST C18 4.6 x 50 mm, 2.5 μm column using ion pair reversed-phase chromatography.1
|
LC system: |
Waters Alliance HPLC Bioseparations System |
|
Column: |
XBridge OST BEH C18 4.6 x 50 mm, 2.5 μm |
|
Column temp: |
60 °C |
|
Flow rate: |
1.0 mL/min |
|
Mobile phase A: |
0.1M TEAA, pH 7.5 |
|
Mobile phase B: |
20% Acetonitrile in A |
|
Gradient: |
30 to 52.5% B in 10.0 min (0.15% ACN/min) |
|
Detection: |
PDA, 260 nm |
|
LC system: |
Waters ACQUITY UPLC System |
|
Column: |
ACQUITY UPLC OST C18 2.1 x 50 mm, 1.7 μm |
|
Column temp.: |
60 °C |
|
Flow rate: |
0.2 mL/min |
|
Mobile phase A: |
0.1M TEAA, pH 7.5 |
|
Mobile phase B: |
20% Acetonitrile in A |
|
Gradient: |
35 to 85% B in 10.0 min (1% ACN/min) |
|
Detection: |
PDA, 260 nm |
Separated products were detected with a Waters PDA detector at 290 nm. The mobile phase A consisted of 0.1 M triethylammonium acetate (TEAA); mobile phase B was 80:20 0.1 M TEAA/acetonitrile. The column temperature was maintained at 60 °C.
As shown in Figure 1, despite the high efficiency of oligonucleotide synthesis, there are many failed sequences present in a 21-mer.
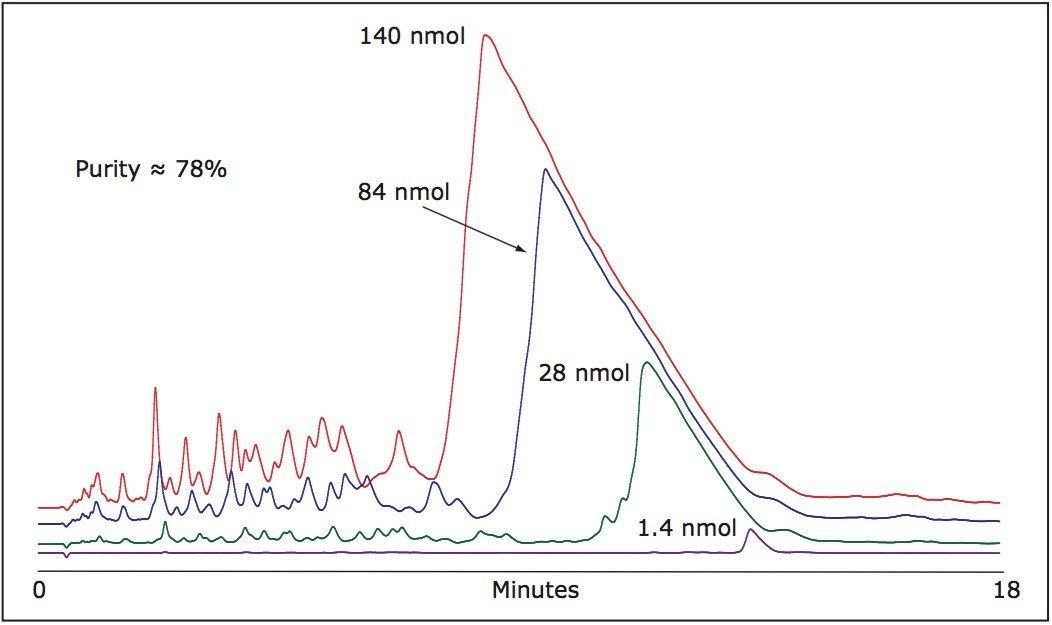
Although the column is overloaded with greater mass loads, the resolution is maintained with N-1, N-2... impurities eluting at the main peak front. Appropriate hearth-cutting of the main 21-mer oligonucleotide peak yields very high purity product.
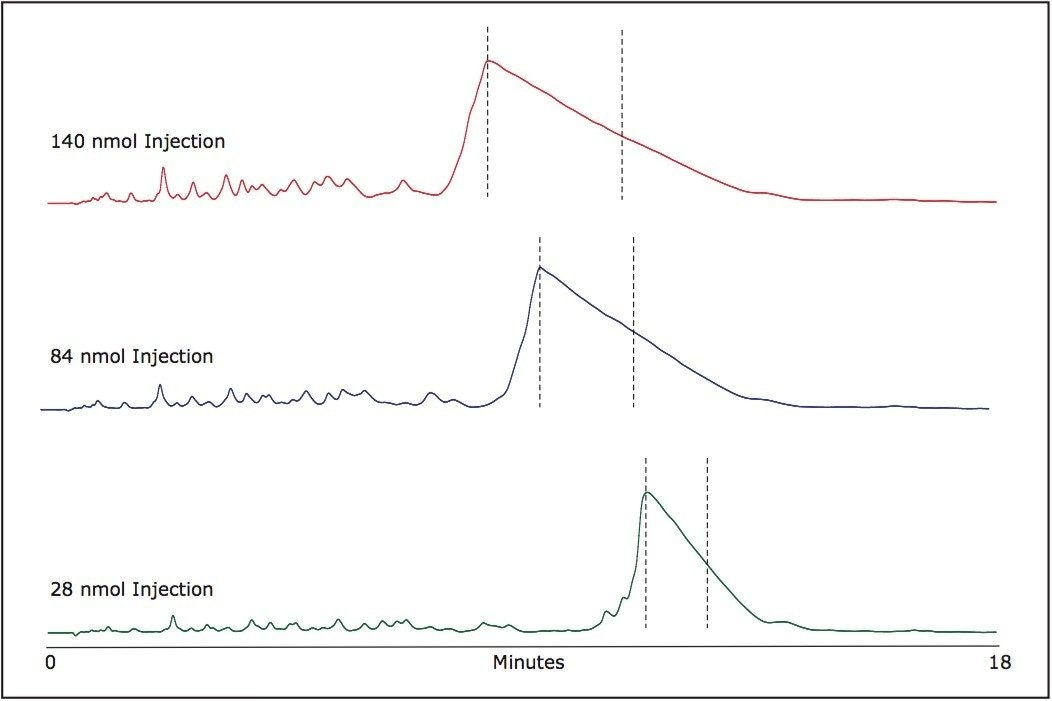
The chosen fraction collection windows are indicated in Figure 2 for various mass loads. Following peak collection, samples can be aliquoted as needed and dried for long term storage. The volatility of TEAA allows for an easy removal of ion-pairing buffer components. The purified oligonucleotides after the solvent evaporation are practically salt free.
The purity of the purified RNA oligonucleotide was verified using the ACQUITY UPLC System. As shown in Figure 3, our purification method efficiently reduces failed sequence impurities and generates an oligonucleotide of superior purity than is available from commercially available oligonucleotides without purification.
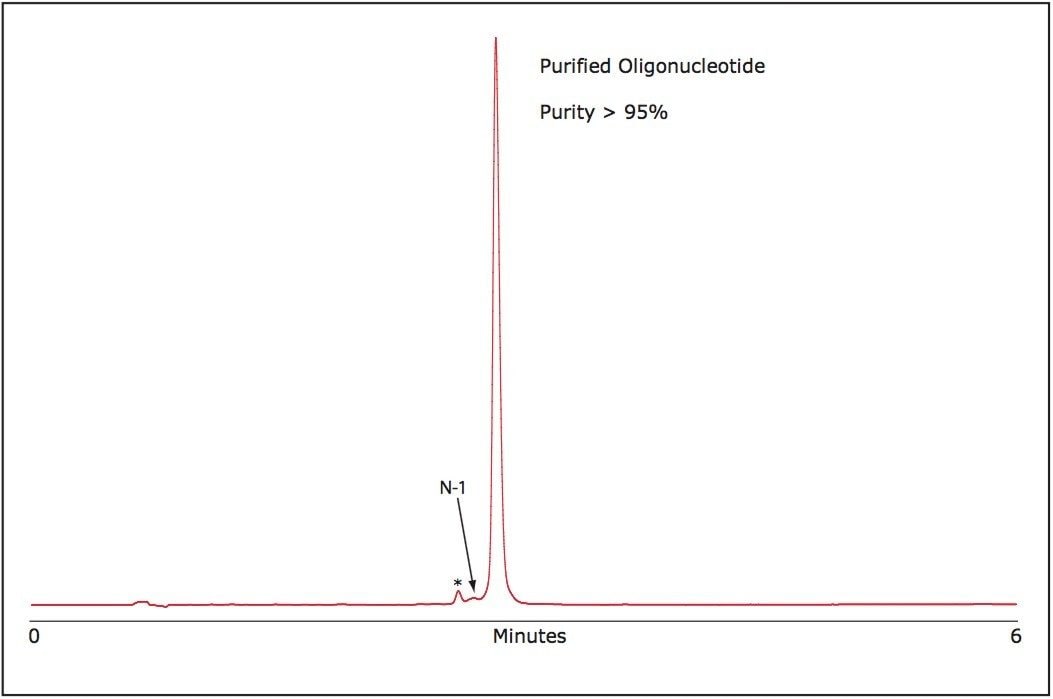
The purification strategy for single-stranded RNA oligonucleotides presented here is rapid, cost effective, and yields high purity material. In a short time, using OST Column chemistry and the Alliance HPLC Bioseparations System, large quantities of crude single-stranded RNA can be successfully purified yielding material of high purity, ca. 95%, with an estimated yield of 55% based on collected peak area to the total peak area of the sample.
This method is particularly useful for the purification of single-stranded RNA for use in RNAi experiments where assurance of purity, and therefore specificity for the target, are of paramount importance.
Additionally, this strategy allows for storage of purified oligonucleotides in the absence of unwanted salts and other impurities often associated with other purification strategies due to the volatile nature of TEAA.
Taken together, this strategy offers a comprehensive purification strategy that is superior to those currently available. Furthermore, this purification method is very cost effective when considering the combined cost of time needed for sample purification, reagents, and the long lifetime of Waters XBridge OST columns.
720002602, April 2008