In this application, we demonstrate how to prepare purified 2-AB labeled N-linked glycans released from glycoproteins such as monoclonal antibodies (IgG1).
Recombinant glycoprotein drugs destined for human use have been widely developed in the biopharmaceutical industry in the last decade. Glycosylation of proteins has a direct impact on their biological activities. Since the nature of protein glycosylation is highly dependent on biosynthesis conditions, monitoring and controlling the biomanufacturing processes of glycotherapeutics is required. New analytical methods for glycan/glycoprotein characterization are highly desirable. Fast and reliable methods for sample cleanup in glycan analysis are an essential part of the biopharmaceutical analytical method toolbox.
Several technologies are currently being used in glycan characterization, such as mass spectrometry (MS), high performance anion-exchange chromatography with pulsed amperometric detection (HPAE-PAD), capillary electrophoresis (CE), and liquid chromatography (LC) with fluorescence (FLR) detection.
In LC-FLR, glycans are typically labeled with 2-aminobenzamide (2-AB), which permit highly-sensitive fluorescence detection.1 This technique allows the quantitation of relative amounts of individual glycans in a heterogeneous complex mixture. In addition, 2-AB labeled glycans provide improved sensitivity in ESI-MS and MALDI-MS for glycan mass profiling analyses.
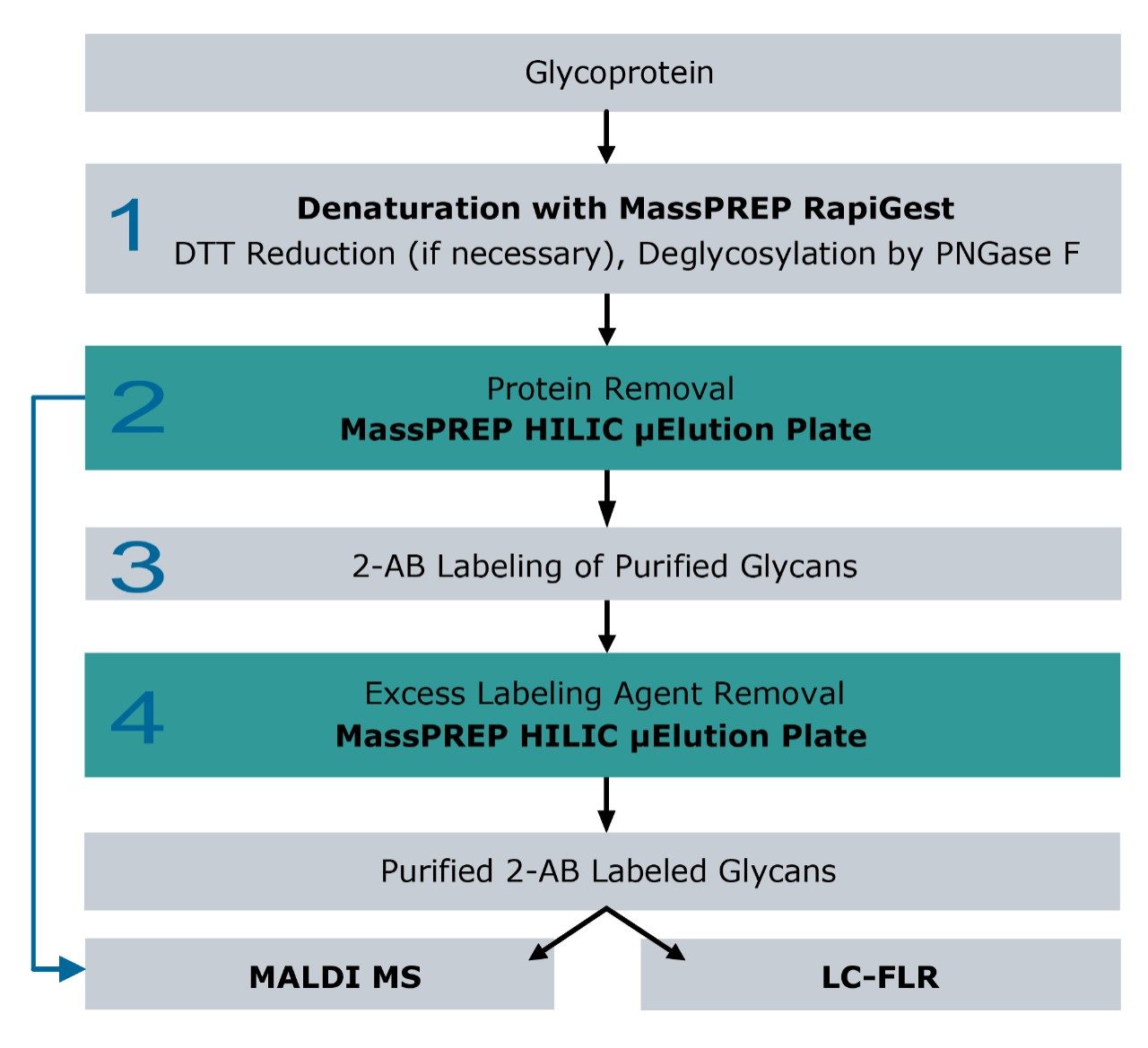
The preparation of purified 2-AB labeled glycans released from glycoproteins can be time consuming, with multiple steps involving deglycosylation using glycosidases and glycan enrichment followed by 2-AB derivatization (Figure 1).
In this application, we demonstrate how to prepare purified 2-AB labeled N-linked glycans released from glycoproteins such as monoclonal antibodies (IgG1). The goal of this method is to provide a rapid, efficient cleanup protocol for LC-FLR, MALDI-MS, and LC-MS analyses using the Waters MassPREP Glycoanalysis Kit.
The MassPREP Kit provides simple and robust sample purification without compromising sample recovery. This complete sample cleanup solution streamlines glycan method development by reducing time and expenses. This kit includes:
Figure 1 schematically presents the workflow for glycan sample preparation and the downstream analyses. The following workflow details each step in the protocol.
Monoclonal mouse IgG1 (VICAM, Division of Waters) and ribonuclease B (Sigma) were prepared separately. Each glycoprotein was solubilized in 0.1 % (w/v) RapiGest SF solution prepared in 50 mM NH4HCO3 buffer at pH 7.9. RapiGest partially denatures the glycoprotein, which exposes the glycosylated site to enzymes for faster and more complete deglycosylation.2
IgG1 was reduced with 20 mM dithiothreitol (DTT) for 45 minutes at 65 °C. DTT reduction was not performed in the ribonuclease B preparation. The enzyme PNGase F (Sigma) was added to the protein solutions and the samples were incubated overnight at 37 °C in sealed vials.
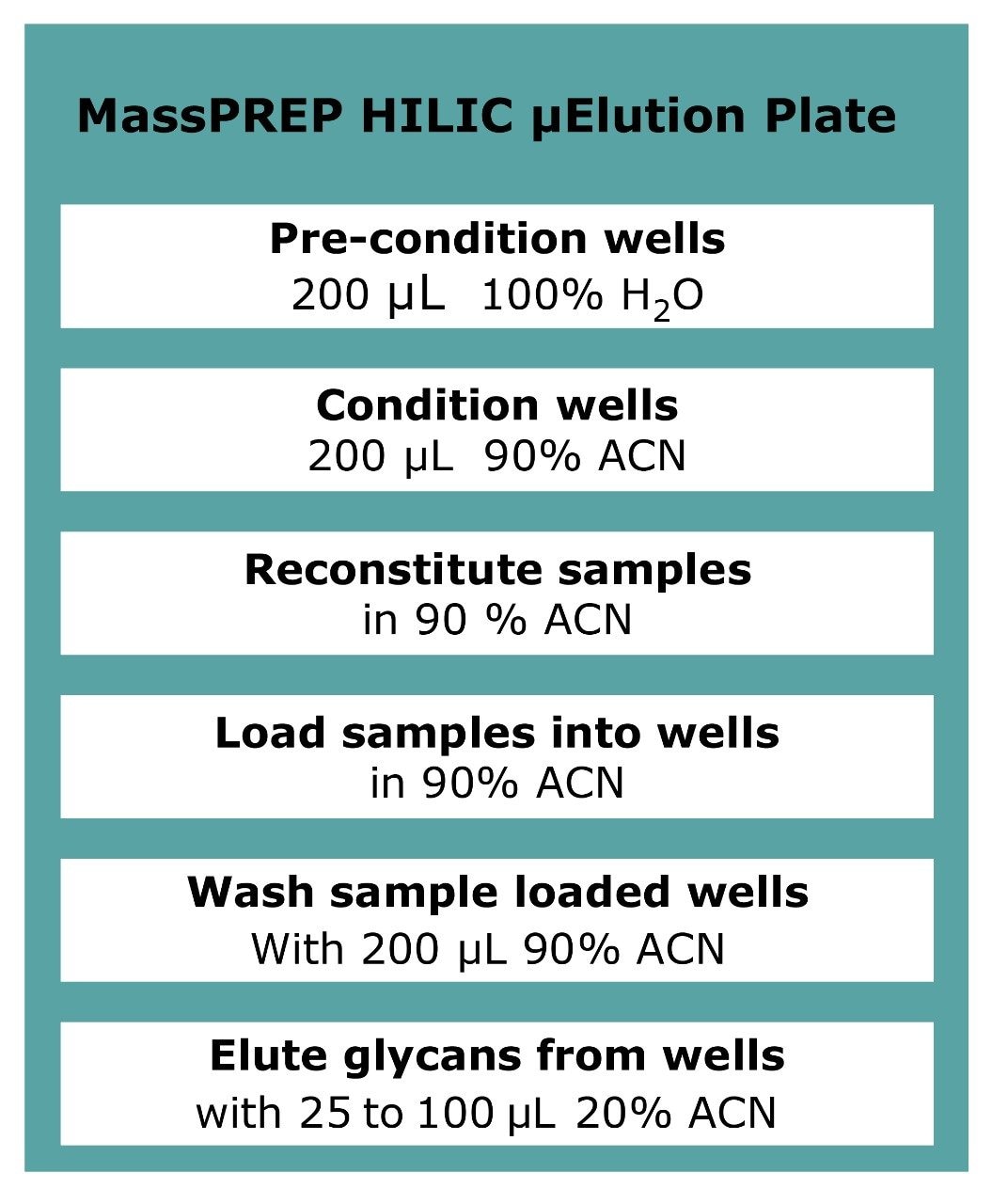
Enzymatically released N-linked glycans were extracted from deglycosylated protein solutions using a MassPREP HILIC 96-well μElution solid-phase extraction (SPE) plate.3
When loaded in high acetonitrile content, HILIC SPE sorbent retains glycans (hydrophilic compounds). More hydrophobic compounds and surfactants are not retained and washed out from the sorbent. The glycans are eluted with solvents containing predominantly water. The MassPREP HILIC plate was utilized following guidelines in Figure 2.4 The eluted glycans were then dried.
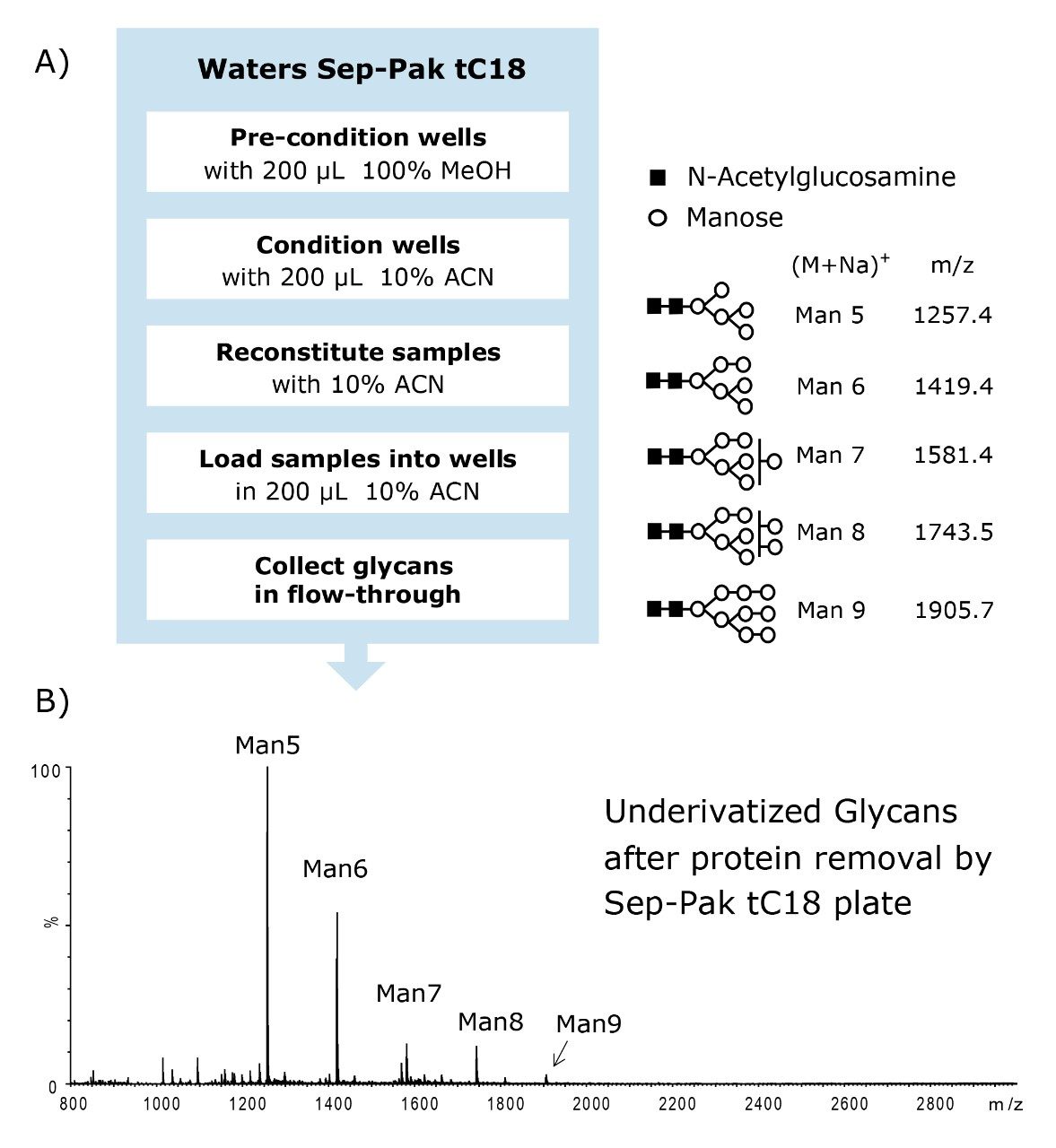
In some cases when a deglycosylated protein solution containing free glycans is reconstituted in 90% acetonitrile, the proteins present in the sample tend to precipitate and restrict flow in the HILIC SPE plate. The Waters Sep-Pak tC18 μElution 96-well plate effectively removes soluble and insoluble proteins from the glycan sample.
The flow-through eluent from the Sep-Pak tC18 protocol, which is shown in Figure 4A, contains free glycans in 10% acetonitrile. The flow-through eluent was dried for use in the labeling process. (Note: the Waters Sep-Pak tC18 μElution 96-well plate can be purchased separately from the MassPREP Glycoanalysis Kit.)
Isolated glycans were labeled using the 2-AB labeling kit from Sigma. 150 μL of acetic acid and 300 μL of dimethyl sulfoxide (DMSO) were mixed, and 100 μL of the acetic acid/DMSO mix was added into 5 mg of 2-AB. The entire volume of 2-AB solution was then added into 6 mg of sodium cyanoborohydride. Five to 10 μL of the final labeling solution was added into dried glycans (100 pmol to 50 nmol) and incubated for 3 hours at 65 °C in a sealed vial.
The 2-AB labeled glycan solution from step 3 was diluted to 90% acetonitrile to reach the HILIC loading condition. The excess labeling reagents were effectively removed from 2-AB labeled glycans using the same HILIC protocol as shown in Figure 2. The purified labeled glycans were then analyzed by MALDI-MS or by LC with FLR detection.
Ultra-pure MassPREP Matrix, DHB, was used in the MALDI-TOF MS analysis. The DHB matrix was reconstituted in 500 μL of pure ethanol to a final concentration of 20 mg/mL. The purified 2-AB labeled glycans were mixed with the DHB matrix at a 1:1 ratio. One μL of the glycan/DHB mix was placed onto a stainless steel MALDI target.
|
MS system: |
Waters MALDI micro MX |
|
Ionization mode: |
Reflectron positive |
|
Pulse voltage: |
1950 v |
|
Detector voltage: |
2350 v |
|
Laser: |
250 |
|
Laser firing rate: |
10 hz |
|
Acquisition range: |
800 to 3000 m/z |
Sample cleanup is critical for both underivatized and labeled glycans in MS analyses. For underivatized glycans, the HILIC SPE plate can be used to remove protein, salt, and surfactant prior to MALDI-MS as shown for N-glycans released from IgG1 (Figure 3A). During labeling, an excess 2-AB is required to achieve complete derivatization of glycans.
However, the excess of labeling reagent often needs to be removed from the sample prior to LC-FLR and MS analyses. A suitable protocol using MassPREP HILIC SPE is shown in Figure 1, Step 4. The method successfully recovers 2-AB labeled glycans as shown in Figure 3B.
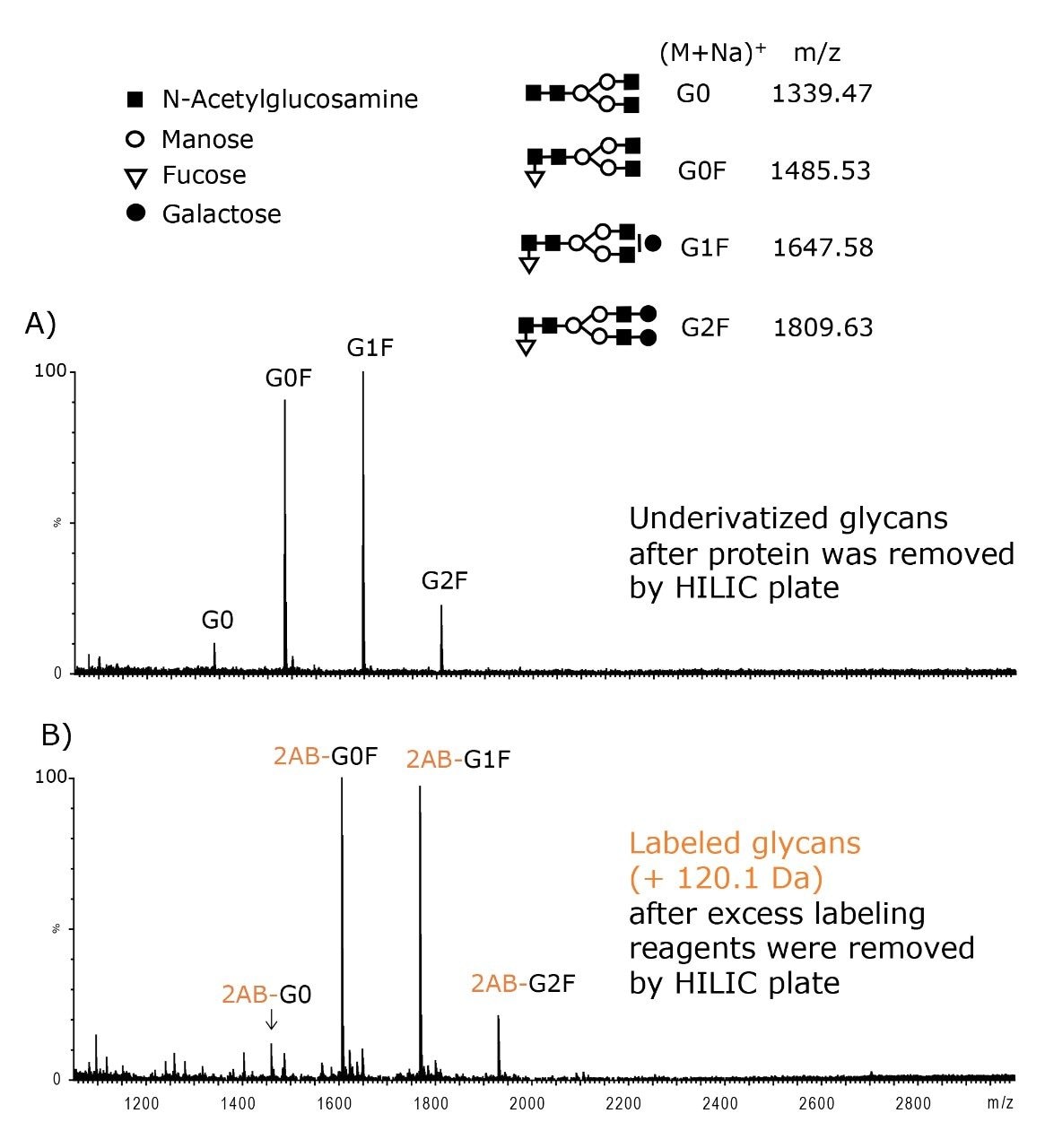
Because the HILIC sorbent has sufficient affinity for hydrophilic glycans, they can be highly enriched on the SPE plate. With sample loads of several hundred microliters, glycans can be recovered in ~25 to 100 μL of eluent. On the contrary, the cleanup method shown in Figure 4B utilizing the Sep-Pak tC18 SPE plate does not allow for sample concentration. Sep-Pak tC18 SPE devices can be used for protein removal, as recommended, but the glycans recovered in the sample flow-through are neither concentrated nor desalted.
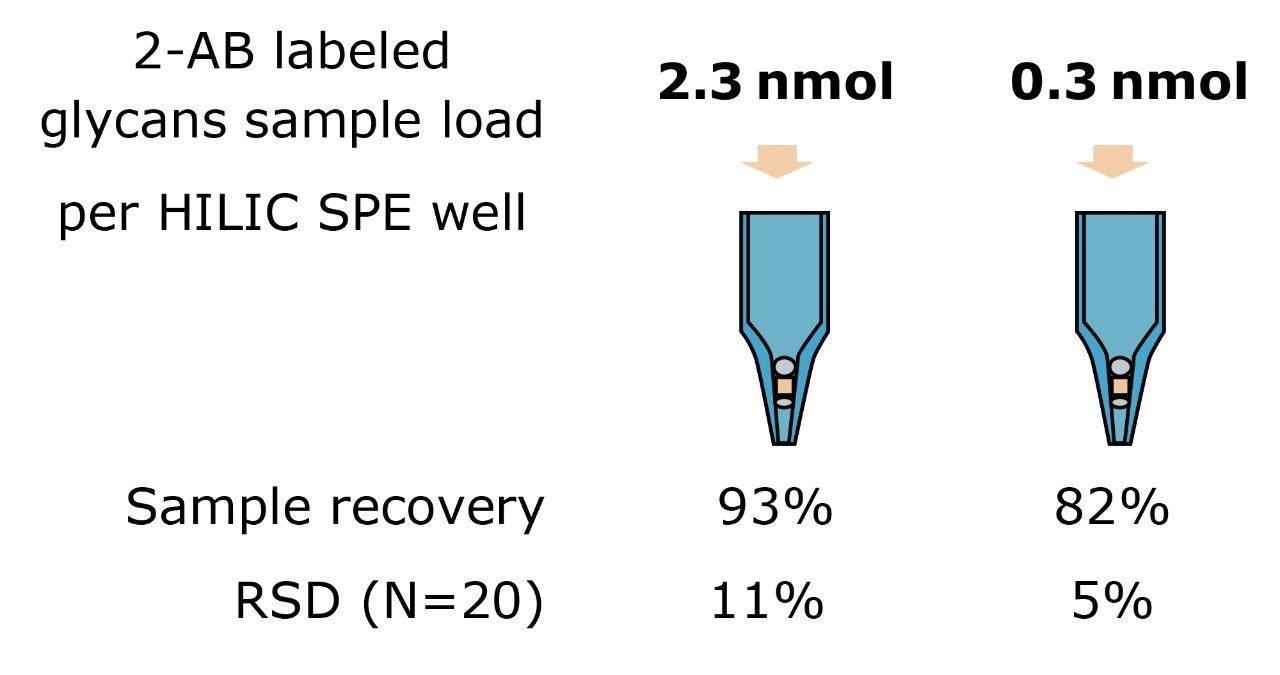
LC-FLR analysis was used to assess the labeled glycans recovery after HILIC cleanup. The FLR signals of 2-AB labeled glycans were compared to the injections of the sample prior to SPE cleanup. Results are summarized in Figure 5. Approximately 0.3 nmol or 2.3 nmol of 2-AB labeled N-glycans released from ribonuclease B were loaded in an HILIC plate and eluted out with 200 μL of 20% acetonitrile. Excellent SPE recovery was achieved for both mass loads with good reproducibility in 20 parallel experiments (Figure 5).
We have shown the use of the MassPREP HILIC μElution plate for extraction of underivatized and 2-AB labeled glycans. The method is fast and reproducible. The key advantages of this method include:
The MassPREP Glycoanalysis Kit is designed for biopharmaceutical applications, minimizing the amount of sample required for the analysis. Because of the format of μElution SPE plate, small volumes of eluents permit shortened evaporation time, eliminating the bottleneck in sample preparation.
Rapid and effective sample preparation helps to improve laboratory productivity and analysis throughput. Increased throughput plays an essential role in reducing the time needed for method development. This, in turn, reduces the amount of materials and laborious processes necessary for validation and qualification of methods developed in biopharmaceutical laboratories.
720002622, June 2008