In this application note E. coli was used as a model organism to demonstrate the capabilities of this system for detailed quantitative and qualitative analysis of a complex biological system.
Many studies involving protein analysis of complex protein mixtures have been accomplished by combining the well-established separation capabilities of two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) with mass spectrometry (MS)-based or tandem mass spectrometry (MS/MS)-based sequence identification of the gel-resolved proteins. Although 2D-PAGE has been used successfully in quantitative proteomics, it is subject to mass range and pI limitations and it requires considerable effort to generate reproducible results. Gel spots may also contain more than one protein which will affect subsequent quantitative analysis.
The subject of this study is an alternative label-free, LC-MS approach which has been developed to enable quantitation and identification of proteins from a single experiment.
The Waters Protein Expression System utilizes the high retention time reproducibility of the Waters nanoACQUITY UPLC System and the exact mass measurement accuracy capability of the Micromass Q-Tof Premier Mass Spectrometer. In addition a novel suite of Informatics tools have been developed, to process and interpret meaningful quantitative/qualitative information from the complex datasets obtained. The Waters Protein Expression System enables the researcher to determine both the changes in relative abundance of peptides across samples and controls, as well as to identify the parent proteins from the same experiment.1-5
The patented LC-MS method presented here employs a multiplexed acquisition routine which enables the parallel analysis of the constituent peptides in a complex biological sample, leading to a significant improvement in the sequence coverage obtained for identified proteins.6
In previous work, the approach used in the Protein Expression System was shown to yield extensive quantitative and qualitative information from a series of protein mixtures in a background of human serum.7 The work presented in this application note was designed to use E. coli as a model organism to demonstrate the capabilities of this system for detailed quantitative and qualitative analysis of a complex biological system.

E. coli was grown in the presence of minimal media and one of three primary carbon sources; Glucose, Lactose or Acetate. The digested, constituent proteins from each of the samples were analyzed in triplicate using the Waters Protein Expression System to determine relative peptide expression changes and to identify the parent proteins.
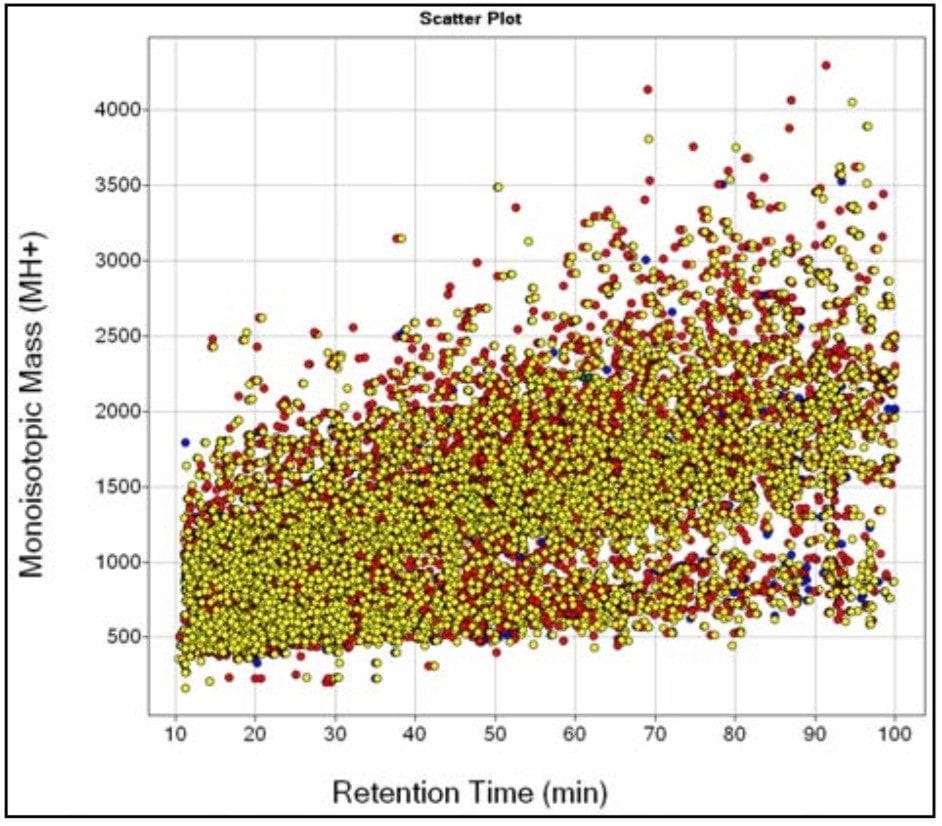
The complexity of the three E. coli samples is demonstrated in Figure 2, where the average monoisotopic mass for each of the extracted peptide components (MH+) is displayed against its corresponding average retention time for all three conditions. This is known as the EMRT, or Exact Mass Retention Time, which is used as a specific signature for a given peptide allowing it to be identified in a sample with high specificity, and tracked across sample sets for subsequent quantitative comparison. The peptide ions (EMRT’s) from replicate injections were clustered by their exact mass and retention time. These clustered EMRT pairs can be plotted to display the up and down regulation of peptides between samples. Subsequent databank searching of selected clusters leads to identification of the protein(s).
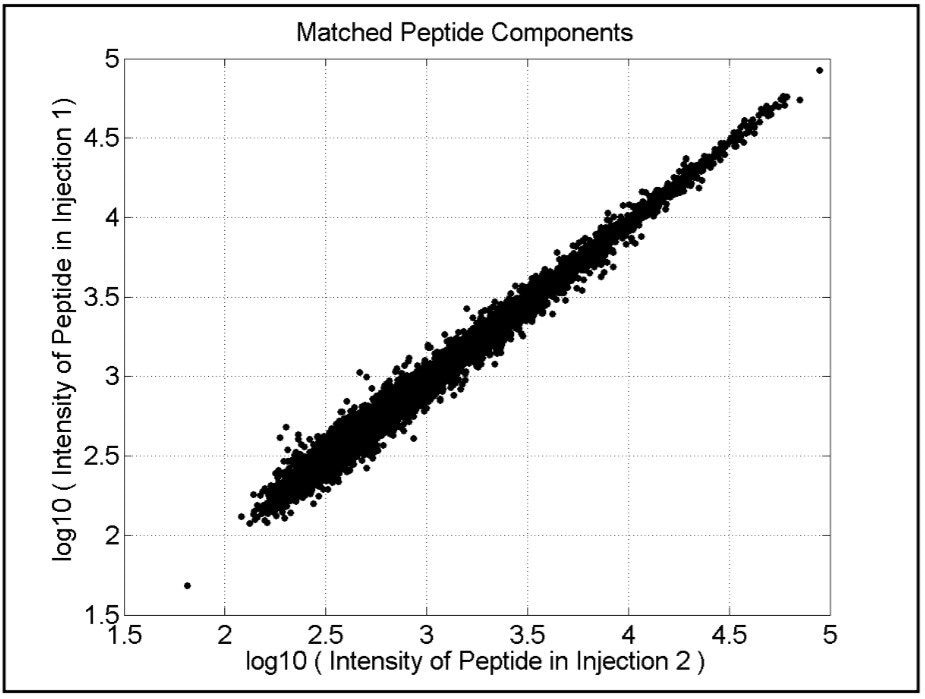
The analytical reproducibility of the method is shown in Figure 3, which compares two repeat injections of the E. coli protein digest sample grown on acetate, demonstrating a coefficient of variation of approximately 13%. Pair-wise comparisons of replicate injections of the E. coli grown on glucose and lactose were 17% and 15%, respectively. This degree of analytical performance ensures discovery of small expression level differences between samples.
The discovery of low-level protein abundance changes is also enabled by the Informatics normalization schemes, auto-normalization of the peptide intensities or normalization to an internal standard. Four peptides from protein chain elongation factor, TUFA, were chosen for global normalization throughout the entire experiment. The four TUFA peptides chosen for normalization were found in all replicate injections for the three conditions. The peptides were identified using databank searching against the elevated energy (MSE) data. Figure 4 shows the annotated MSE mass spectrum for the AIDKPFLLPIEDVFSISGR peptide (2117.1479 MH+ at 91.81 min) from TUFA. Peptides were also identified from succinyl-CoA synthetase, isocitrate lyase and citrate synthase (data not shown).
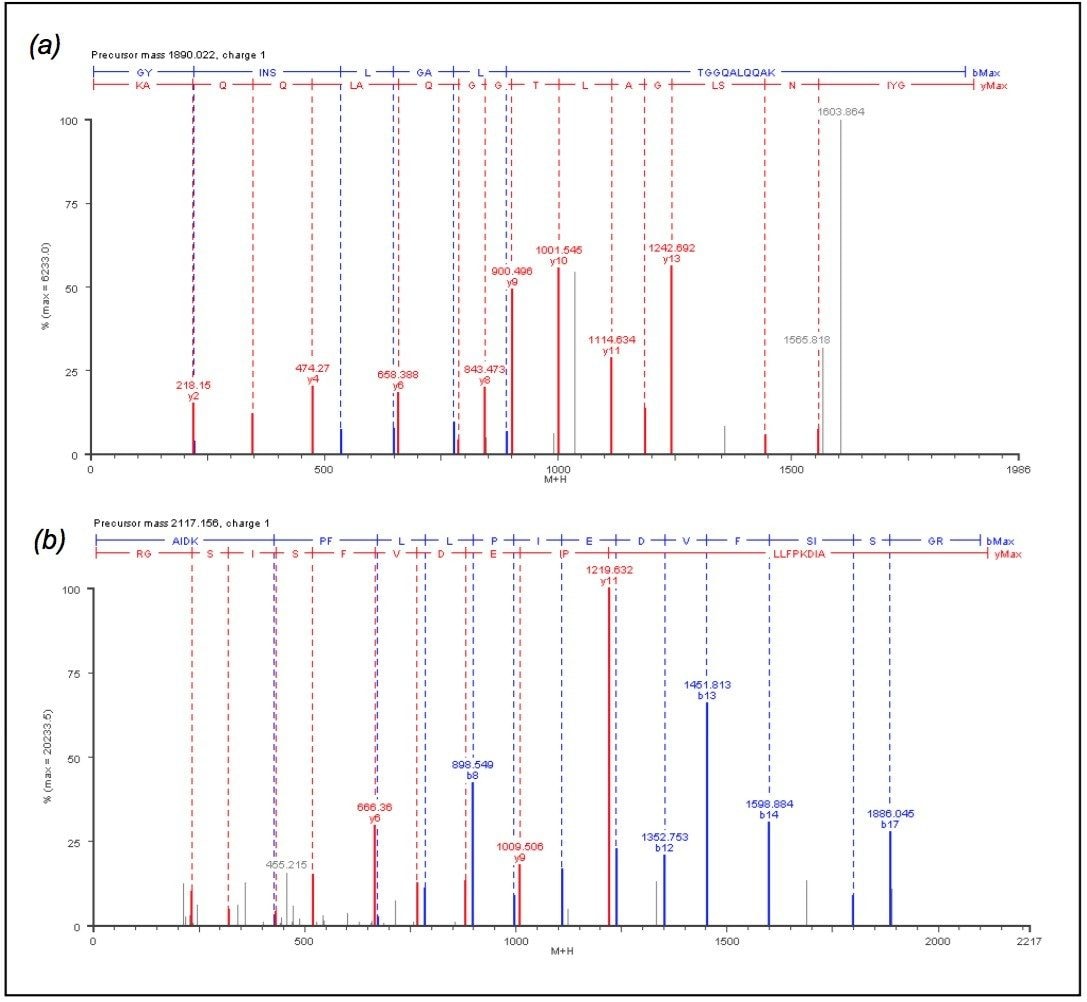
Figure 5a displays the relative peptide abundance observed in the glucose versus lactose growth conditions. Once the matched peptides are plotted according to their relative fold-change, the quantitative comparison of the matched peptides provides a means to quickly identify those specific peptides/proteins that exhibit a noticeable change due to the perturbation.
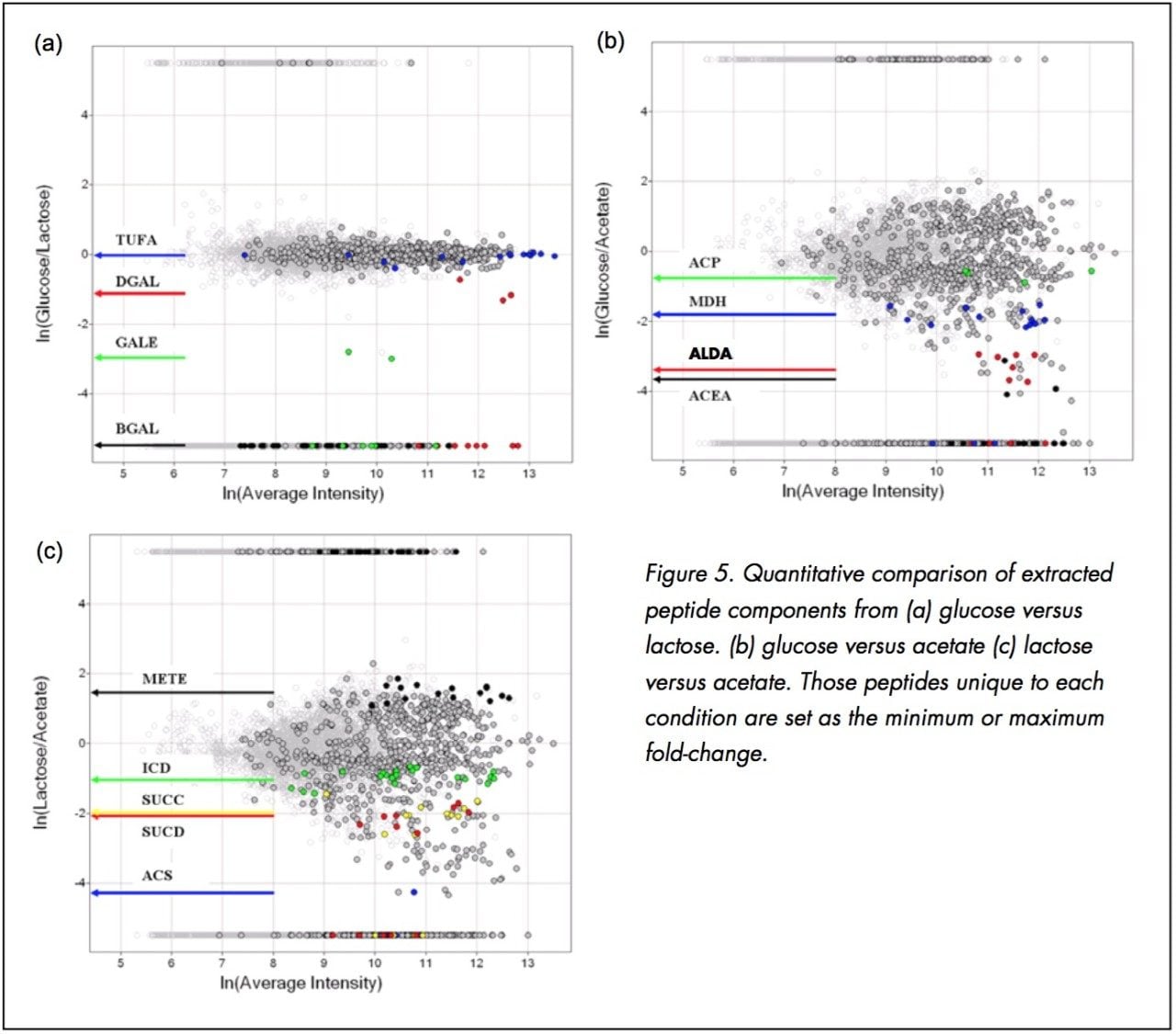
The comparison in Figure 5a represents the natural log (ln) of the average intensity for the matched peptide components across both conditions (x-axis) versus the ln of the normalized intensity ratio for the matched peptides between the two conditions (y-axis).
Those peptides which were unique to each condition were set to either 5.5 (unique to glucose) or -5.5 (unique to lactose).
The average protein coverage obtained from the individual injections for TUFA was approximately 55%. The inherent redundancy of the tryptic peptides to any particular protein provides multiple independent quantitative measurements and can be used to determine the relative quantitation of any particular protein between two conditions. The multiple measurements also provide a means to determine a confidence interval for the relative quantitation of any particular protein in a study.
Protein chain elongation factor, TUFA, was found in all three conditions and was determined to not be differentially expressed (Table 1). Galactose-binding transport protein (DGAL), UDP-galactose-4-epimerase (GALE), beta-D-galactosidase (BGAL) were also found to be differentially expressed (Figure 5a).
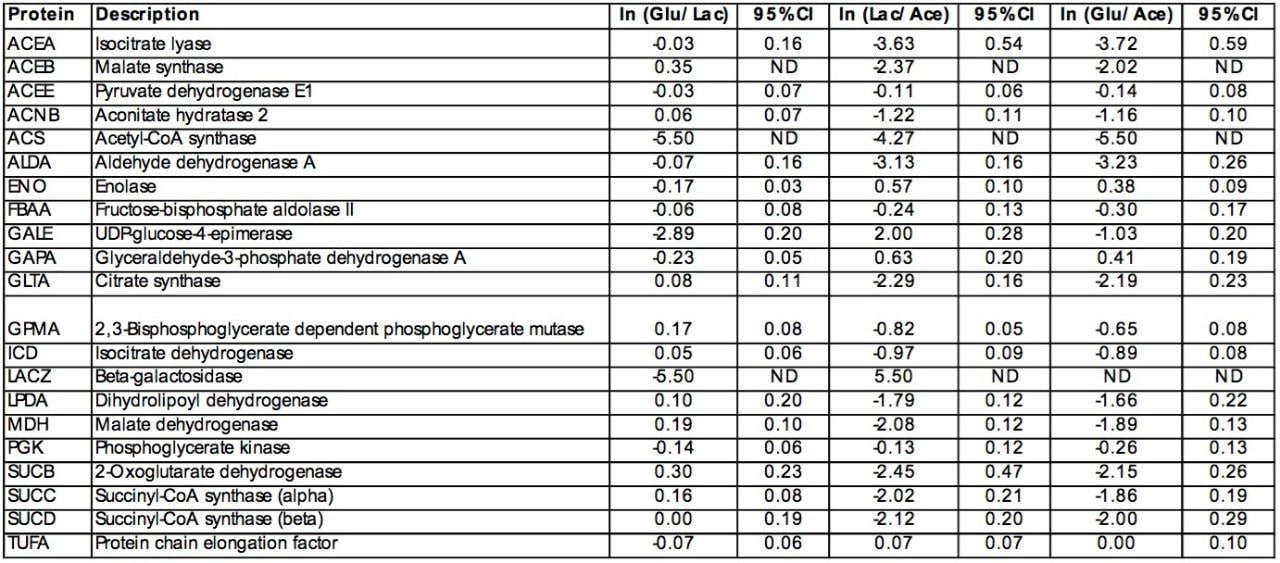
Figure 5b and Figure 5c show the differential peptide analysis between those peptides found in the comparison of glucose/acetate and lactose/acetate, respectively. Highlighted in Figure 5b are the peptides identified to acyl-carrier protein (ACP), malate dehydrogenase (MDH), aldehyde dehydrogenase A (ALDA) and isocitrate lyase (ICL or ACEA) from the quantitative comparison of the peptides found in the glucose versus acetate growth conditions.
Figure 5c illustrates the peptides identified to tetrahydropteroyl-triglutamate-homocysteine methyltransferase (METE), Isocitrate dehydrogenase (ICD or IDH), Succinyl-CoA-synthetase (SUCC), Succinyl-CoA-synthetase (SUCD) and Acetyl-CoA synthase (ACS) from the quantitative comparison of the peptides found in the lactose versus acetate growth conditions. Interestingly, SUCC and SUCD interact to form the heterotetrameric A2B2 complex of succinyl-CoA synthetase. The observed fold-change for SUCC and SUCD is consistent with the structure.
The limited differential expression observed between lactose and glucose can be explained by the metabolic requirements for the utilizing the two different carbon sources. Lactose is a disaccharide of glucose and galactose, and therefore the metabolic differences are manifested in the active transport of the disaccharide carbon source and the conversion and epimerization of the galactose monomer to glucose. Figure 5a highlights the peptides identified to the galactose-specific processing proteins: DGAL, GALE and BGAL. It is not surprising that there is little variation associated with the observed peptide components in Figure 5a, since there is minimal impact to the downstream metabolic activity when supporting growth on either glucose versus lactose. Both carbon sources are ultimately processed through the glycolysis and the citric acid cycle in a similar fashion.
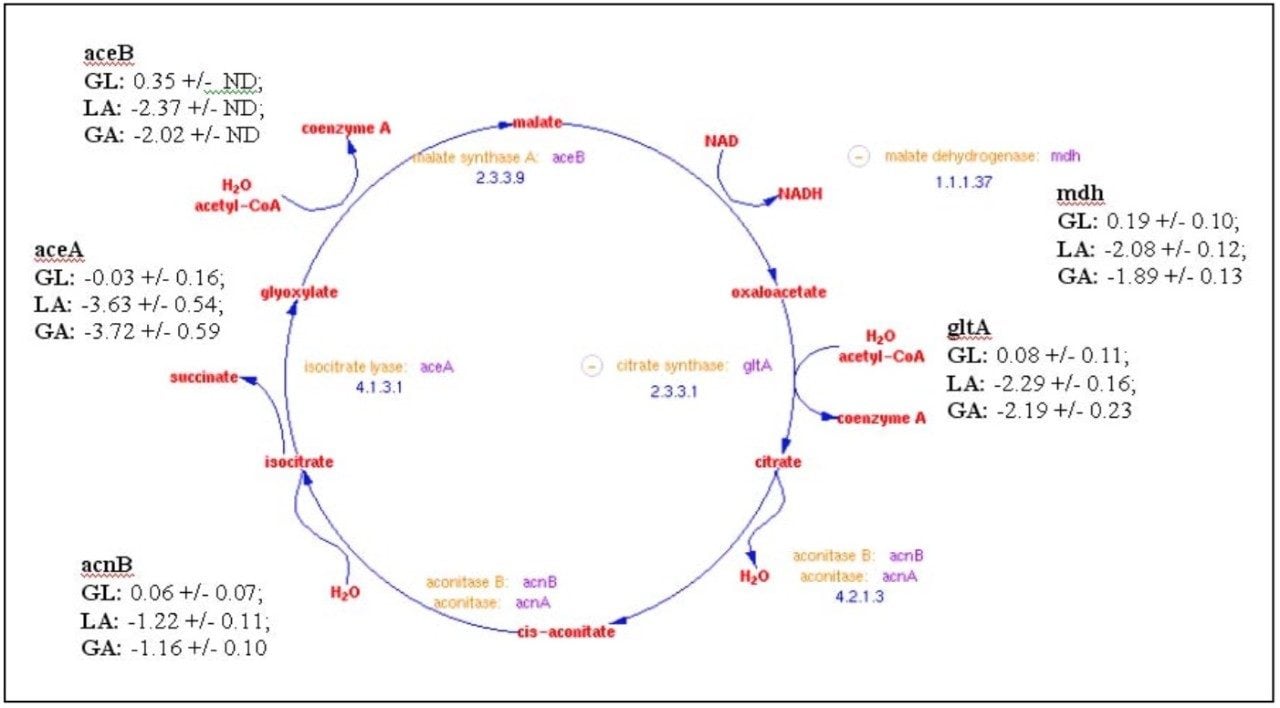
The relative fold-change of a few of characterized proteins from each binary comparison is illustrated in Table 1. Providing acetate as the sole carbon source for E. coli requires substantially different metabolic activity to support growth than that of either glucose or lactose. This is evident from the variation associated with the relative foldchange associated with the binary comparisons of the detected peptide components from either glucose or lactose to those of acetate (Figure 5b and 5c). Acetate is a simple carbon source which initially bypasses glycolysis and enters into a modified version of the citric acid cycle, glyoxylate shunt (Figure 6), to provide the necessary primary metabolites and energy to support growth. Before entering the glyoxylate shunt, acetate must first be converted to acetyl-CoA. The conversion of acetate to acetyl-CoA is performed through the activity of acetyl-CoA synthetase (ACS). The evidence accumulated from the peptide analysis of the three conditions indicates that ACS was not detected in the glucose condition, but was present in both the lactose and acetate conditions. The relative quantitation obtained from the comparison of lactose or glucose versus acetate is consistent with the induction of ACS in the acetate condition. Another indication of growth on acetate as the sole carbon source is the activation of the glyoxylate shunt which is accompanied with the relative induction of isocitrate lyase (ACEA) and malate synthase (ACEB) as seen in Table 1.
Interestingly, over 60% of the ribosomal proteins as well as a few translation factors were identified from the three different growth conditions. The average protein coverage obtained from these abundant proteins was approximately 40%. The quantitation results indicated that the level of the ribosomal machinery in acetate was approximately 2.5 times lower than the levels observed in either glucose or lactose. These results are in agreement with the consequences to E. coli when subjected to poor growth conditions.8-10
6. Bateman, R. H; Hoyes, J. B., U. K. Patent 2, 364, 168 A, 2002.
7. Silva, J. C.; Denny, R.; Dorschel, C.; Gorenstein, M.; Kass, I. J.; Li, G. Z.; McKenna, T.; Nold, M. J.; Richardson, K.; Young, P. and Geromanos, S. (2005) Analytical Chemistry, April 1; 77(7): 2187–2200.
8. Marr, A. G. (1991) Microbiology Reviews; 55(2):316–333.
9. Toa, H.; Bausch, C.; Richmond, C.; Blattner, F. R. and Conway T. (1999) J. Bacteriol. 181(20):6425–6440.
10. Silva, J. C.; Denny, R.; Dorschel, C.; Li, G. Z.; Richardson, K.; Wall, D. and Geromanos, S. Simultaneous Qualitative and Quantitative Analysis of the E. coli Proteome: A Sweet Tale“. Manuscript in Preparation.
11. Hughes, M.; C.; Silva, J. C.; Dorschel, C.; Geromanos, S. and Townsend, C. A. In Proceedings of the 53nd Annual ASMS Conference on Mass Spectrometry and Allied Topics, “Target Identification Studies: Application of a New LC-MS and Data Analysis Software System to Identify Drug-Induced Changes in Mycobacteria”, 2005, San Antonio, TX, USA.
720001310, August 2005