For research use only. Not for use in diagnostic procedures.
An analytically sensitive, accurate and precise UPLC-MS/MS method has been developed for the analysis of mycophenolic acid in human plasma for clinical research purposes.
This method provides separation of mycophenolic acid from its metabolites through the use of selective chromatography with no significant carryover and minimal matrix effects.
Mycophenolic acid is an immunosuppressant agent that acts by inhibiting inosine monophosphate dehydrogenase in the purine synthesis pathway required for the growth of B and T cells. Mycophenolic acid is metabolised into mycophenolic acid glucuronide (MPAG) and mycophenolic acid acyl-glucuronide (AcMPAG).
Several existing methods for the analysis of mycophenolic acid lack the ability to separate mycophenolic acid from its metabolites MPAG and AcMPAG. Both glucuronide metabolites undergo in-source fragmentation to generate an interference in the mycophenolic acid MRM transition. In this method, UPLC-MS/MS is able to chromatographically separate mycophenolic acid from its metabolites in a run time of 3 minutes.
A protein precipitation method for the extraction of mycophenolic acid from plasma for clinical research has been developed. Chromatographic separation was performed on an ACQUITY UPLC I-Class System using an ACQUITY UPLC HSS C18 SB Column followed by detection on a Xevo TQD Mass Spectrometer (Figure 1).

Sample preparation
Calibrators were prepared using ClinCal multi-level calibrator set (Recipe, Munich, Germany). In-house calibrators were prepared to extend the linear range by spiking mycophenolic acid (Cerilliant, Round Rock, TX, USA) into pooled human plasma (Sera Laboratories, West Sussex, UK). QC samples were ClinChek multi-level control set (Recipe, Munich, Germany). Tri-deuterated (2H3) mycophenolic acid (Cerilliant, Round Rock, TX, USA) was used as the internal standard at a concentration of 0.2 μg/mL in 30% aqueous methanol containing 0.1 M zinc sulfate (precipitating solution).
Sample extraction
An aliquot of sample, 50 μL, was transferred to a micro-centrifuge tube, and precipitating solution, 500 μL, was added to each sample.
The samples were capped and vortexed for 20 seconds prior to centrifugation for 5 minutes at 18800 g. The supernatants were transferred to a 96-well 1 mL plate and sealed for analysis.
|
System: |
ACQUITY UPLC I-Class with FTN |
|
Column: |
ACQUITY HSS C18 SB, 2.1 x 30 mm, 1.8 μm (P/N 186004117) |
|
Mobile phase A: |
Water with 2 mM ammonium acetate and 0.1% formic acid |
|
Mobile phase B: |
Methanol with 2 mM ammonium acetate and 0.1% formic acid |
|
Wash solvent: |
90% aqueous methanol and 0.1% formic acid |
|
Purge solvent: |
30% aqueous methanol |
|
Seal wash: |
20% aqueous methanol |
|
Pre-inject wash time: |
0 seconds |
|
Post-inject wash time: |
30 seconds |
|
Column temp.: |
50 °C |
|
Sample temp.: |
4 °C |
|
Injection volume: |
10 μL |
|
Flow rate: |
0.70 mL/min |
|
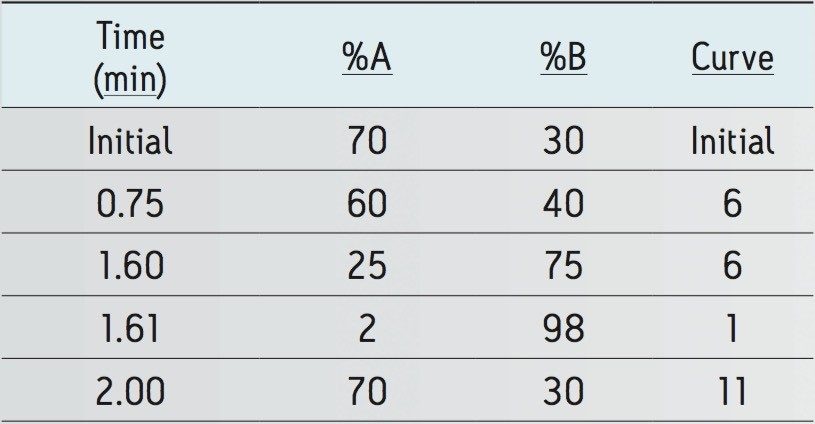
Gradient: |
Table 1 |
|
Run time: |
2.5 minutes (approximately 3.0 minutes injection to injection) |
|
System: |
Xevo TQD |
|
Resolution: |
MS1 and MS2 (0.75 FWHM) |
|
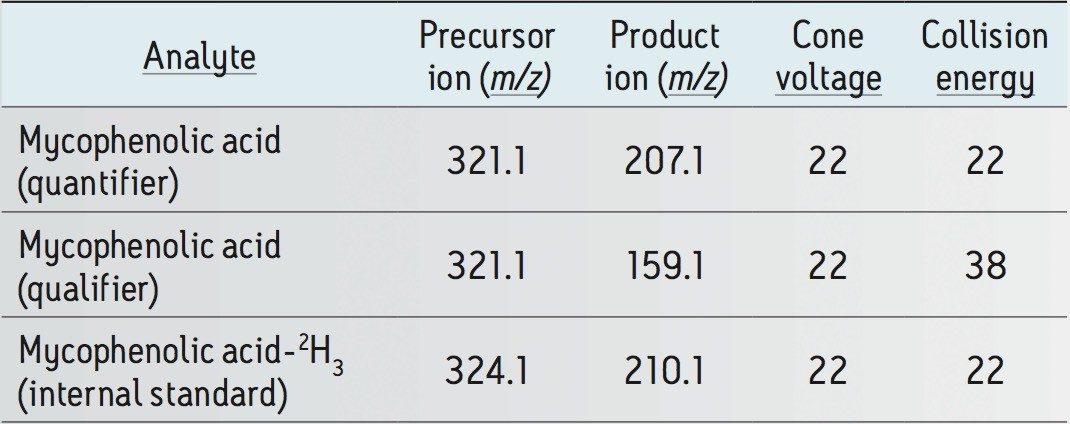
Acquisition mode: |
Multiple reaction monitoring (MRM) (Table 2) |
|
Polarity: |
ESI positive ionization |
|
Capillary voltage: |
3.5 kV |
|
Cone voltage: |
Table 2 |
|
Source temp.: |
150 °C |
|
Desolvation temp.: |
600 °C |
|
Dwell time: |
0.05 seconds |
|
Inter-scan delay: |
0.02 seconds |
|
Inter-channel delay: |
0.01 seconds |
|
Data management |
|
|
MassLynx Software v4.1 with TargetLynx Application Manager |
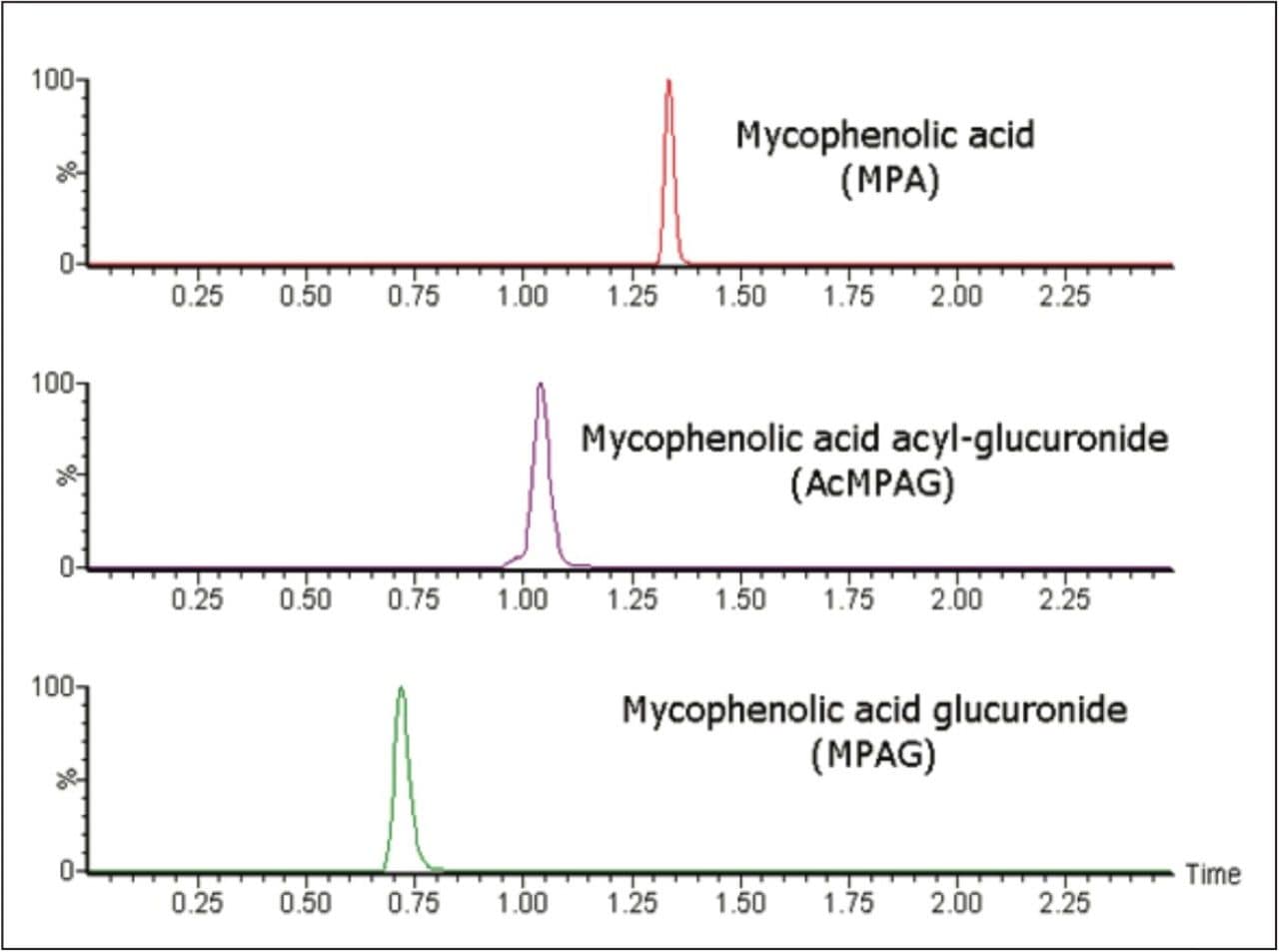
The method was shown to be linear over the range of 0.1–20.0 μg/mL when different ratios of high and low concentration pools of mycophenolic acid were combined and analysed in replicates of four. Calibration lines were linear with coefficient of determinations (r2) >0.994. Chromatographic separation of the metabolites of mycophenolic acid was achieved (see Figure 2).
Analytical sensitivity investigations demonstrate that the method would allow precise quantification (<20% RSD) at 0.075 μg/mL. The signal-to-noise ratio of the 0.075 μg/mL samples was >10:1 for ten replicates over three days.
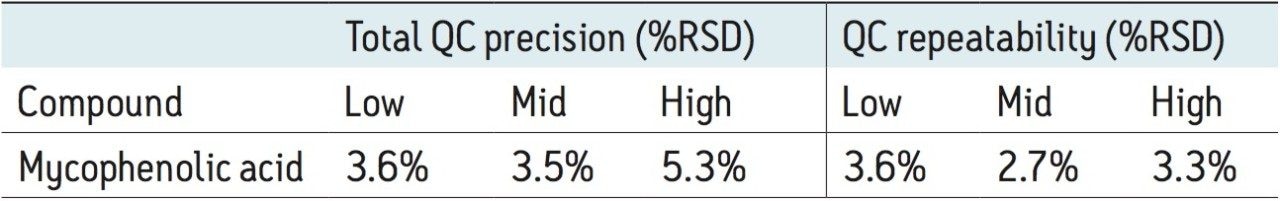
Precision was determined by extracting and quantifying five replicates of tri-level QC material once per day over five separate days (n=25). The results of these experiments are shown in Table 3, where total precision and repeatability at the low (0.5 μg/mL), mid (2.4 μg/mL), and high (5.0 μg/mL) concentrations is ≤5.3% RSD.
No significant system carryover was observed from high concentration samples prepared at 40 μg/mL, into subsequent blank injections. A 1:1 dilution was successfully performed on an over-range sample prepared at 40 μg/mL, providing a mean accuracy of 98% with a %RSD of 5.9%.
Matrix effects were described by matrix factor with and without internal standard. Mean matrix effects were 0.91 and 0.94 for low and high QC concentrations respectively. Calculations using analyte:internal standard peak area response ratio for low and high QC concentrations gave mean matrix effects of 0.97 and 0.96, respectively. These values demonstrate that the internal standard is compensating for minor ion suppression when the analyte alone is examined.
Samples (n=12) were obtained from the International Proficiency Testing (IPT) Mycophenolate Scheme (Bioanalytics.co.uk) and were analysed with the obtained values compared to the HPLC/MS method mean. The determined bias was ≤5.1%.

The bias observed between control and test samples (spiked with high level of interference) for the quantitation of mycophenolic acid was <10% for all interferences tested. Interference testing included the metabolites of mycophenolic acid, endogenous compounds including cholesterol and creatinine, and exogenous compounds including itraconazole and everolimus.
A comparison of sample values (n=35) was performed against an independent LC-MS/MS method for the analysis of mycophenolic acid. The data was processed using Analyse-it software (Analyse-it Software Ltd.) and the comparison produced a Deming regression of y=0.90x + 0.13 (Figure 3A). Altman-Bland analysis demonstrated a mean negative bias of -0.05% (Figure 3B).

An analytically sensitive, accurate and precise UPLC-MS/MS method has been developed for the analysis of mycophenolic acid in human plasma for clinical research purposes.
This method provides separation of mycophenolic acid from its metabolites through the use of selective chromatography with no significant carryover and minimal matrix effects.
Waters acknowledges Gary Chusney and his colleagues at the Leslie Brent Laboratory, Hammersmith Hospital, London, UK, for the provision of anonymized human plasma samples and for the valued assistance and advice used to produce this application note.
720005463, July 2015