This is an Application Brief and does not contain a detailed Experimental section.
This application brief demonstrates the benefits of Waters Xevo G2-S QTof Mass Spectrometer and UNIFI Scientific Information System to obtain a deeper understanding of metabolite identification samples for the comprehensive detection, confident identification, and interrogation of metabolite profiles.
Ultimate benchtop exact mass MS performance meets next-generation software processing to achieve the most comprehensive and accurate metabolite identification studies.
Workflows continue to be optimized throughout the drug development process. Even small gains when multiplied over the course of many thousands of samples can result in tangible and significant financial benefits. When performing metabolite identification, the proven UPLC-MSE method facilitates routine detection of major metabolites. The ability to interrogate complex samples and obtain definitive fragmentation profiles using this method allows for both the confirmation of the precursor as true metabolites, as well as the ability to perform confident structural elucidation, which is essential to the process.
Technology improvements incorporated into the Xevo G2-S QTof have resulted in a significant boost in sensitivity via the proven and innovative StepWave ion source design. Combining this with Waters’ next-generation platform – the UNIFI Scientific Information System – allows users to quickly and easily interrogate their samples.
This technology brief gives scientists the tools to comprehensively understand the metabolism of their candidate molecules and to effectively report the information back to their colleagues, customers, or collaborators.
In this application brief we discuss how the Xevo G2-S QTof can benefit biotransformation studies by generating better quality product ion spectra and then reviewing the data in UNIFI Software.
Rat liver microsomes spiked with 2-μM nefazodone were incubated for 0 and 60 min at 37 °C. The samples were quenched with one volume of cold acetonitrile + 0.1% formic acid and centrifuged. The samples were then analyzed using a Waters Xevo G2-S QTof, coupled with a Waters ACQUITY UPLC I-Class liquid chromatography system. Data acquisition was performed using MSE (an acquisition technique that acquires both precursor and product ion information for virtually every detectable component of a mixture) in positive ion, resolution mode (> 32,500 FWHM). 5 μL of sample was injected onto an ACQUITY UPLC BEH, 1.7 μm, 2.1 x 50 mm Column and run with a 5 min. gradient using a flow rate of 0.5 mL/min. The mobile phase consisted of water + 0.1% formic acid (A) and acetonitrile + 0.1% formic acid (B). Data were processed and analyzed using UNIFI Software.
At the core of UNIFI is a scientific library that allows users to map and navigate their metabolite identification data in novel, visually interactive ways, as well as in tabular or combined views. Figure 1 shows the summary view, which enables users to quickly scan through large lists of metabolites more effectively. Comprehensive filtering tools and customizable views allow users to have the precise information they need right at their fingertips.
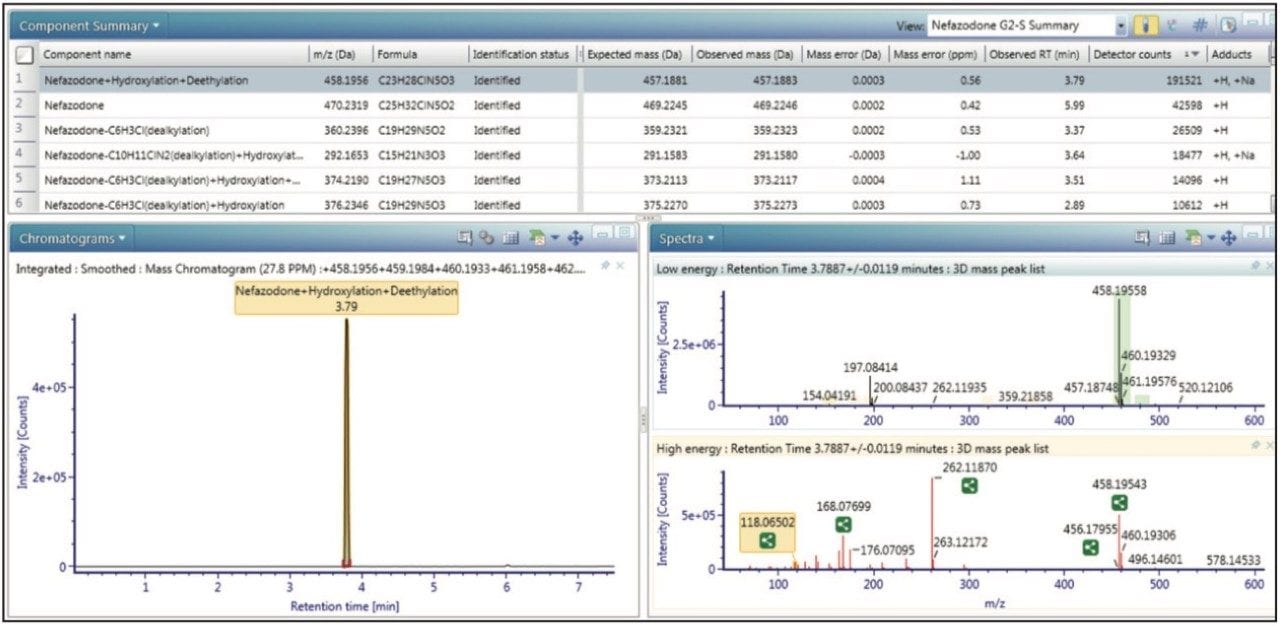
In Figure 2 shows the spectral review of a major metabolite of nefazodone. From the data we can see that excellent mass accuracy was achieved across the mass range. Also shown is the reviewed data annotated with assignments of fragment peaks, including, in this case, both structural and mass spectral information.
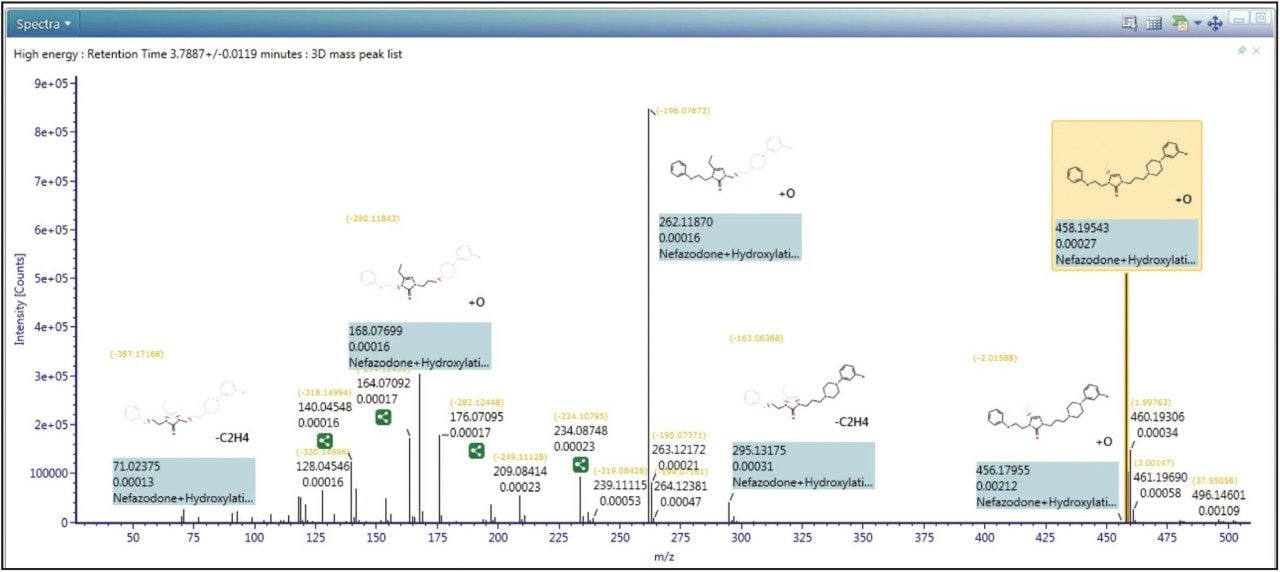
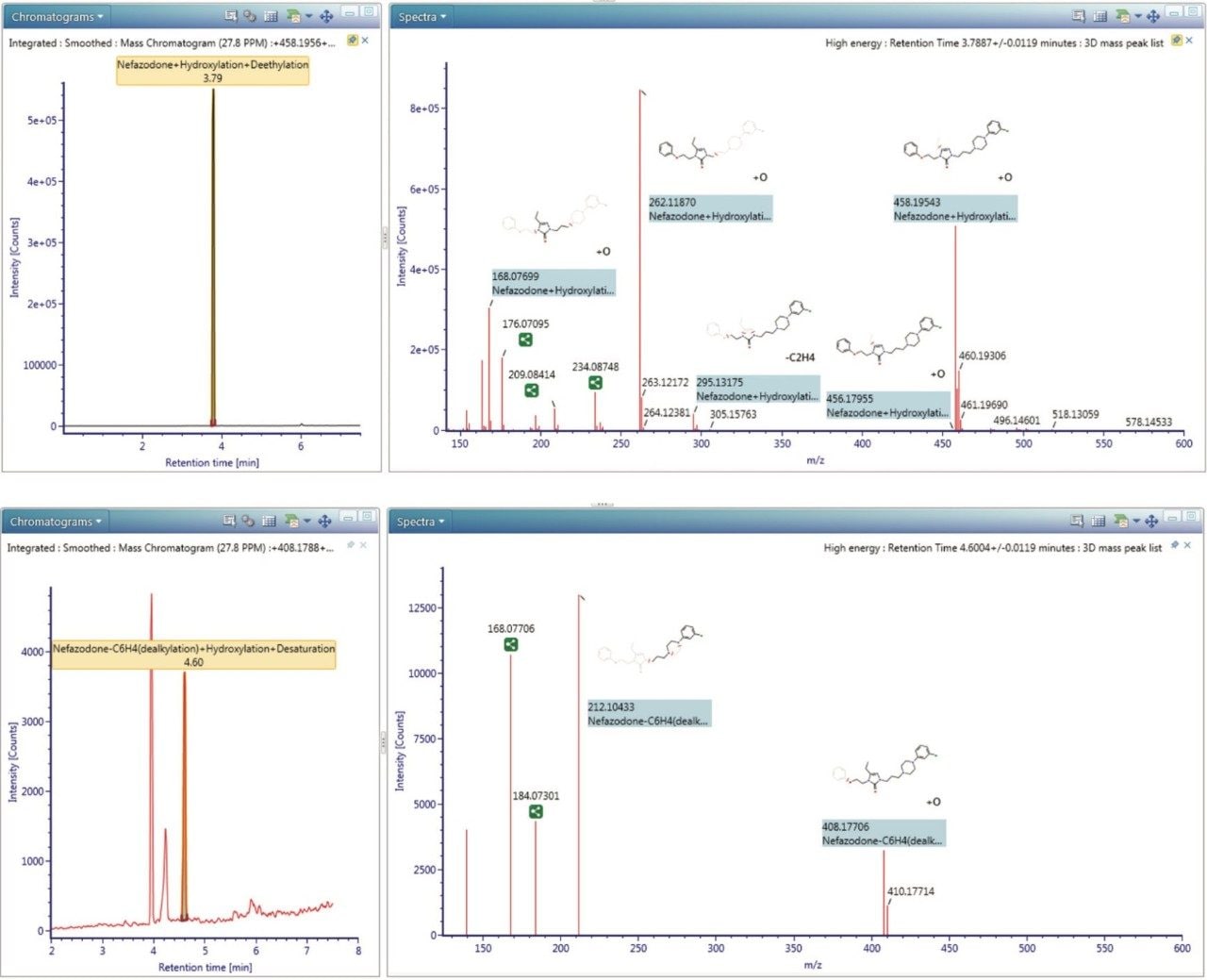
Figure 3 shows the data-rich information captured by the Xevo G2-S QTof, processing information from a complex matrix over a large dynamic range allowing users to identify all of a drug’s metabolites more quickly and comprehensively.
The advances presented in this application brief allow analysts to identify metabolites at lower levels than ever before and interrogate them with innovative, next-generation software.
Xevo G2-S QTof, coupled with the ACQUITY UPLC I-Class System and UNIFI Software provides the comprehensive benchtop UPLC-MSE performance, chemically intelligent informatics, and flexible data management tools to perform complete characterization of drug metabolism samples, ensuring that the results generated can be presented with complete confidence.
720004324, May 2012