This Application note demonstrates the productivity gains possible with the Xevo TQ-S for use in bioanalytical method development.
Where traditionally this activity requires several analyses to collect the necessary full scan, MRM, and precursor ion data to capture information on the analyte peak phospholipids and other endogenous compounds, Xevo TQ-S employs unique RADAR functionality to simultaneously collect these data in the time scale of a narrow 1- to 2-second UPLC peak.
For a bioanalysis method to be effective it must be transferable, reproducible, and robust to variations in matrix caused by diet, phenotype, age, and gender that can cause assay variability. The development of a reliable LC-MS/MS bioanalysis assay involves the optimization of sample preparation, MS detection, and chromatography conditions. This can be a time-consuming process requiring the analyte peaks(s) to be resolved from resolved endogenous matrix components that cause ion suppression and assay irreproducibility. This often requires multiple analytical runs to acquire the necessary multiple reaction monitoring (MRM), product ion, and full-scan data.
|
LC system: |
ACQUITY UPLC |
|
Column: |
ACQUITY UPLC BEH C18 2.1 x 50 mm |
|
Mobile phase: |
A: 0.1% Formic acid or 0.1% aqueous ammonium hydroxide B: Methanol or acetonitrile Gradient 5 to 95% organic over 0 to 2 min |
|
Flow rate: |
600 μL/min |
|
MS system: |
Xevo TQ-S Positive ion electrospray |
|
MRM: |
315 => 129 |
|
Full scan MS: |
50 to 500 mz at 5000 amu/sec |
|
Product ion scan: |
Precursors of m/z = 184 from 200 to 600 |
Endogenous compounds in serum/plasma, in particular phospholipids, can cause ion suppression, leading to reduced sensitivity and reduced robustness. The key to a successful bioanalytical method is to adjust the chromatographic conditions such that the analyte peak is positioned away from the endogenous materials in the sample such as phospholipids, amino acids, and nucleosides. As the concentration and nature of these endogenous materials can vary according to subject age, diet, state of health, and phenotype it is critical that as much robustness is built into the method at an early stage to support the methods use in the later stages of drug development and clinical trials.
Traditional method development activity usually requires several analyses to collect the necessary full scan, MRM, and precursor ion data to capture information on the analyte peak phospholipids and other endogenous compounds; this can significantly reduce productivity. This problem is resolved with the Xevo TQ-S by using its unique RADAR functionality, which enables the simultaneous collection of full scan, MRM, and precursor ion scanning data, all in the time scale of a narrow 1- to 2-second UPLC peak.
Previously we have described the use of low and high pH mobile phases to simplify methods development.2 To illustrate the use of the RADAR technology, a common H2 receptor antagonist ranitidine hydrochloride was spiked into rat plasma, precipitated with acetonitrile (2:1) and analyzed using an ACQUITY UPLC System coupled Xevo TQ-S( Figure 1). The data shown in Figure 2 illustrates the chromatogram obtained when a conventional acidic aqueous buffer and acetonitrile organic modifier gradient is employed, and when a basic aqueous buffer is used.
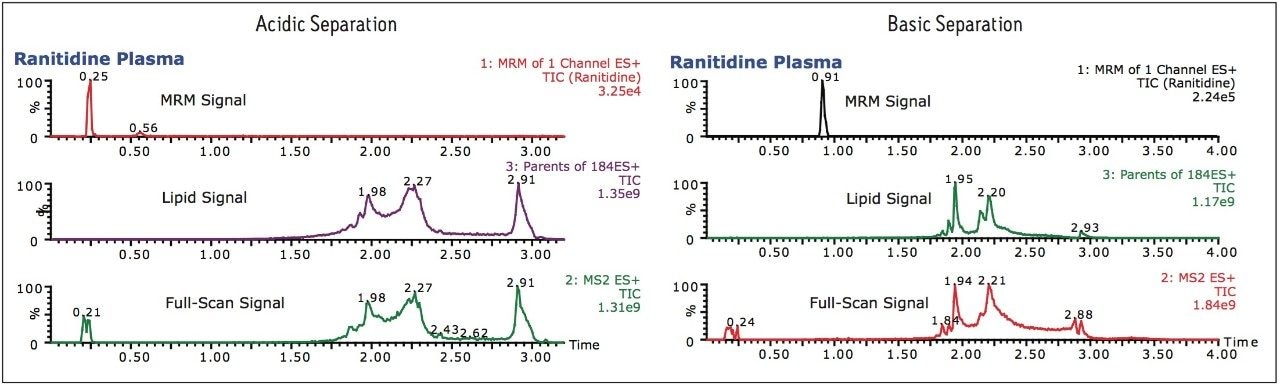
The RADAR MRM data shows that, with an acidic aqueous modifier, ranitidine is not retained, eluting with the void of chromatography system, whereas when a basic aqueous modifier is employed, the compound is retained, eluting at 0.9 min and thus making it the best option. The full-scan and parents of m/z 184 data show that with both the acidic and basic separation the analyte is resolved from phospholipid and other endogenous materials in the matrix.
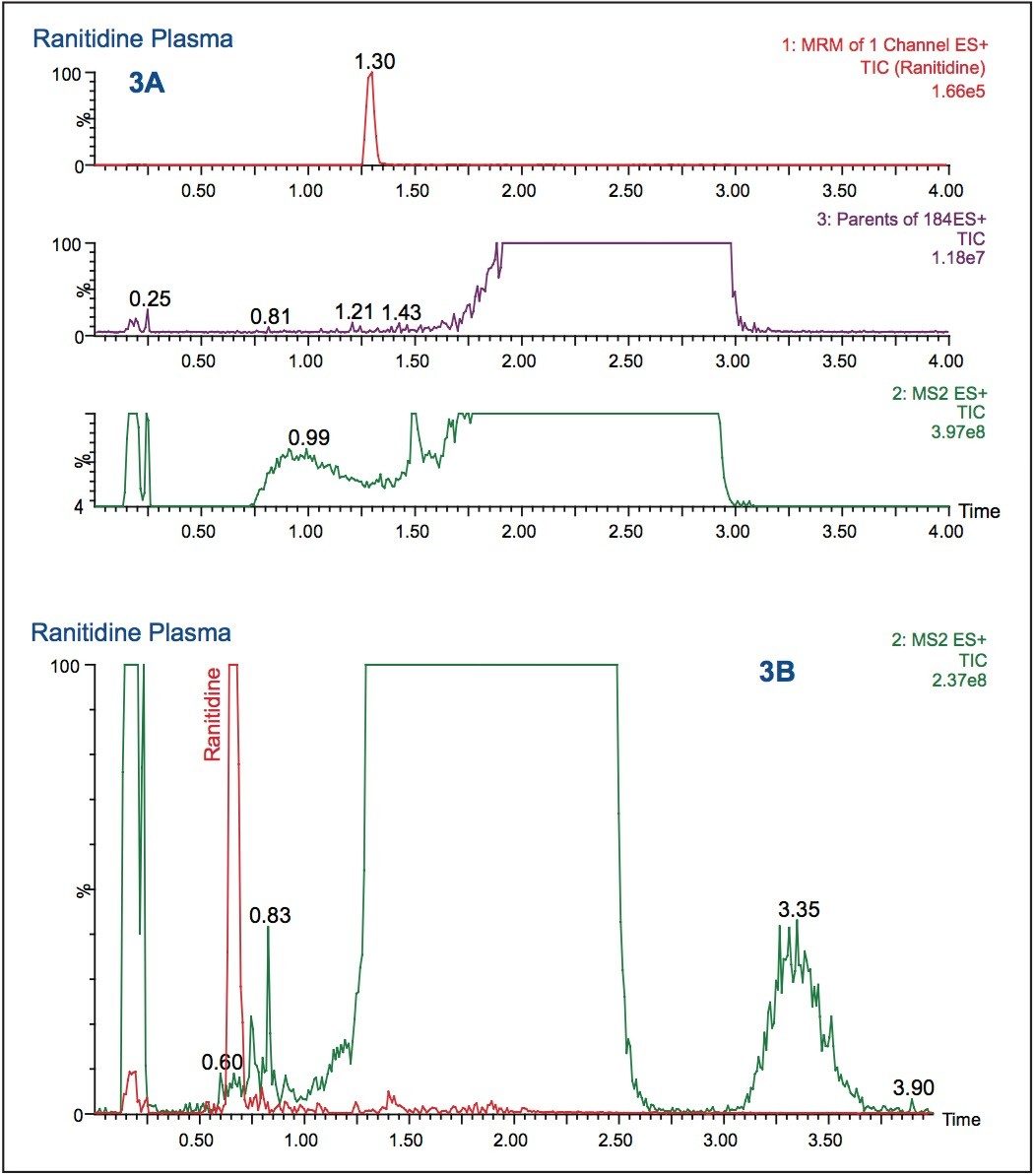
The use of a methanol organic modifier, instead of acetonitrile, with a basic aqueous buffer increased the analyte retention to 1.30 minutes, as shown in Figure 3A. The analyte signal response was also increased by a factor of 4. However, with a simple 5 to 95% basic/ methanol gradient, the full-scan data revealed that the analyte peak coeluted with the endogenous material in the sample. This coelution could cause ion suppression and assay irreproducibility. To resolve the analyte peak from the endogenous material, the gradient steepness was adjusted. The RADAR functionality was used to quickly select the best LC conditions with the best throughput.
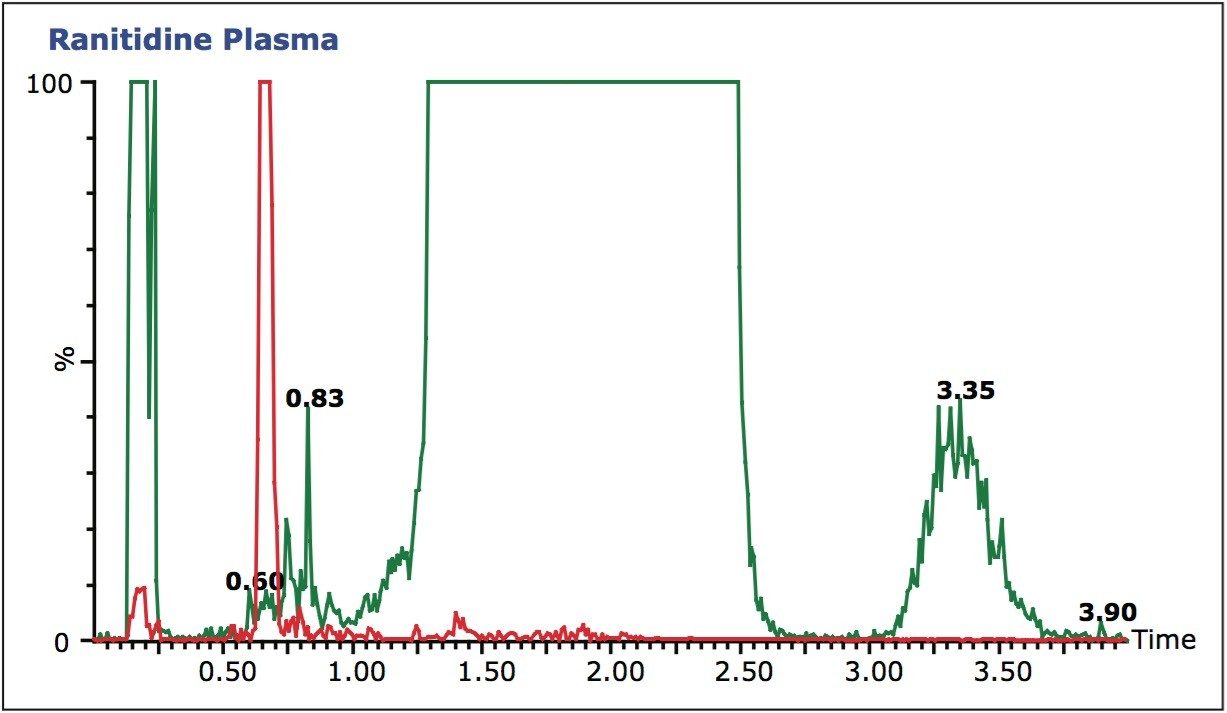
The data displayed in Figure 3B shows the final chromatography conditions. A gradient of 20 to 65% methanol over 1.5 minutes was employed with a 95% organic wash at 1.5 to 2 minutes. As can be seen from the full-scan trace (green), in Figure 3B the analyte peak is well resolved from the endogenous material in the sample. This approach, of simultaneous full-scan MS and MRM data collection, can also be used during sample analysis to check for drug-related metabolites, co-administered therapies, and variations in matrix that could affect the veracity of the results.
Bioanalytical method development is significantly simplified using the Xevo TQ-S with RADAR technology. The ability to simultaneously acquire full-scan data as well as MS/MS and MRM data allows the endogenous sample matrix to be monitored at the same time as the analyte peak. Thus the Xevo TQ-S facilitates:
720003417, January 2011