Peptide Mapping of an Antibody-Drug Conjugate Using PeptideWorks Tryptic Protein Digestion Kit
본 응용 개요서는 구체적인 실험 내용을 포함하지 않습니다.
Abstract
To demonstrate the use of PeptideWorks Tryptic Protein Digestion Kits for Antibody-Drug Conjugate (ADC) peptide mapping.
Benefits
Introduction
Peptide mapping is a powerful tool for protein sequence identification and confirmation, as well as protein modification monitoring, but the procedure is often tedious. It has been shown previously that Waters PeptideWorks Tryptic Protein Digestion Kits (p/n: 176005310) provide an automatable sample preparation procedure and enable reproducible tryptic peptide maps for monoclonal antibodies (mAbs) in less than 2.5 hours.1 In this Technology Brief, we show that insightful information can be obtained from ADC peptide mapping using PeptideWorks Tryptic Protein Digestion Kits.
Experimental
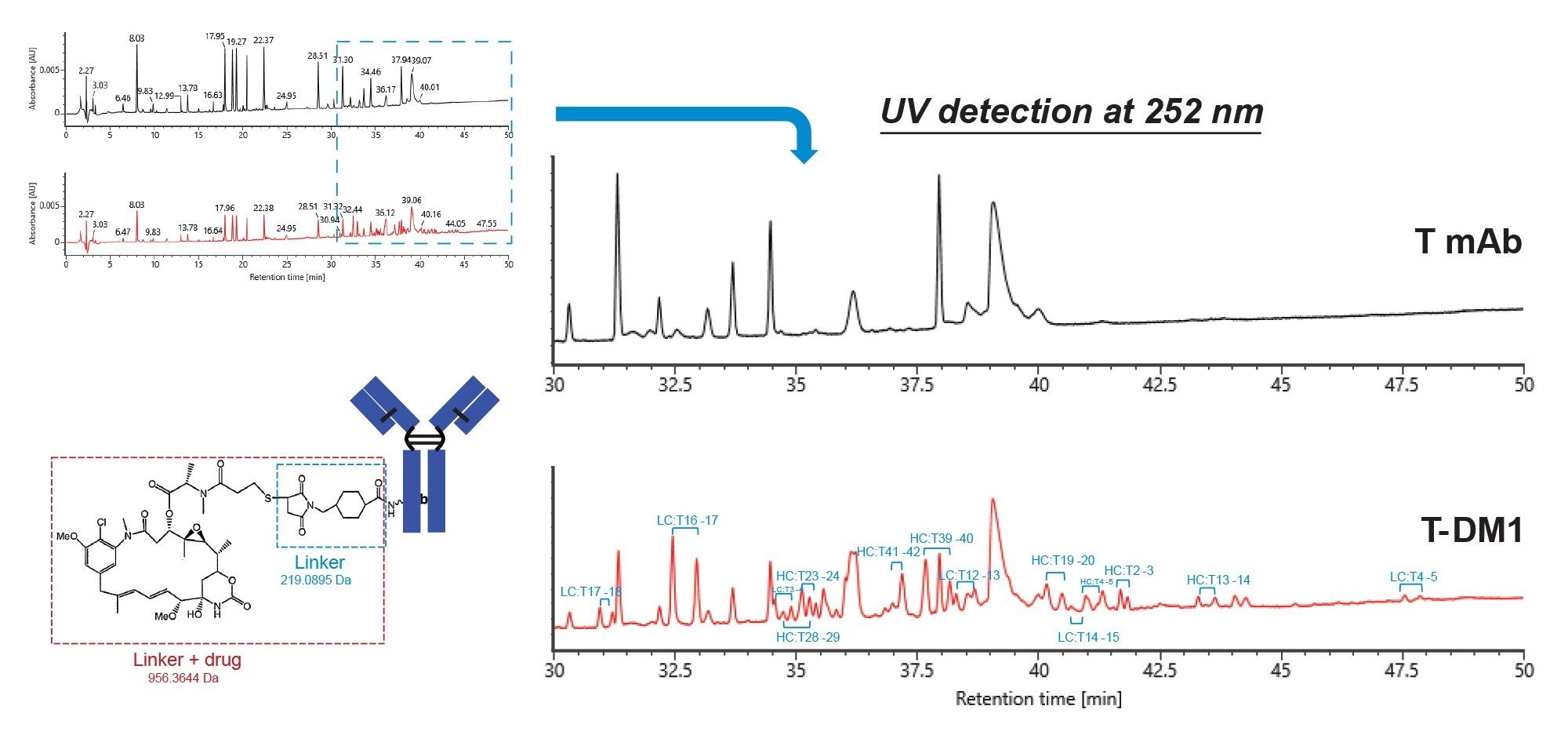
Figure 1 shows UV chromatograms of the peptide maps of both trastuzumab (T mAb) and trastuzumab-emtansine (T-DM1, the ADC). The UV detection was done at 252 nm because the small molecule drug absorbs strongly at this wavelength. A number of additional peaks are observed for the T-DM1 sample in the more retained region of the chromatogram that do not appear for T mAb (the blue dashed box and the blown-up figure). These peaks are likely the drug-conjugated peptides because they elute later on the reversed-phase column due to the hydrophobicity of the small molecule drug. Indeed, many of these additional peaks were confirmed to be the drug-conjugated peptides by mass spectrometry data processed by waters_connect software. As the drug is conjugated through lysine residues, trypsin (Rapizyme Trypsin, MS grade, p/n: 186010106) is unable to digest at the drug-conjugated lysine site, leading to missed cleavage between the lysine-containing peptide, and its following peptide. In addition, these peaks elute in pairs (diastereomers) due to two possible stereochemical configurations during conjugation reaction. About a dozen of them are labeled in Figure 1.
As an example, Figure 2a shows mass spectrometry data of a drug-conjugated peptide-light chain T16–17 (ADYEKHK) peptide. Based on the molecular weight of the peptide and the drug, m/z of 616.27 (+3 charge) was extracted from the BPI chromatograms. As predicted, two diastereomer peaks appeared in the extracted ion chromatogram (XIC) for the T-DM1 sample while no peaks with meaningful information appeared for the T mAb sample. Additionally, the combined spectra of the diastereomer peaks both showed m/z of 616.27 (+3 charge) and 923.90 (+2 charge) as the highest intensity, indicating that the major component of the peaks is the drug-conjugated light chain T16–17 peptide. The diastereomers also appear on the UV chromatogram for the T-DM1 sample but not for the T mAb sample, shown on Figure 2b.
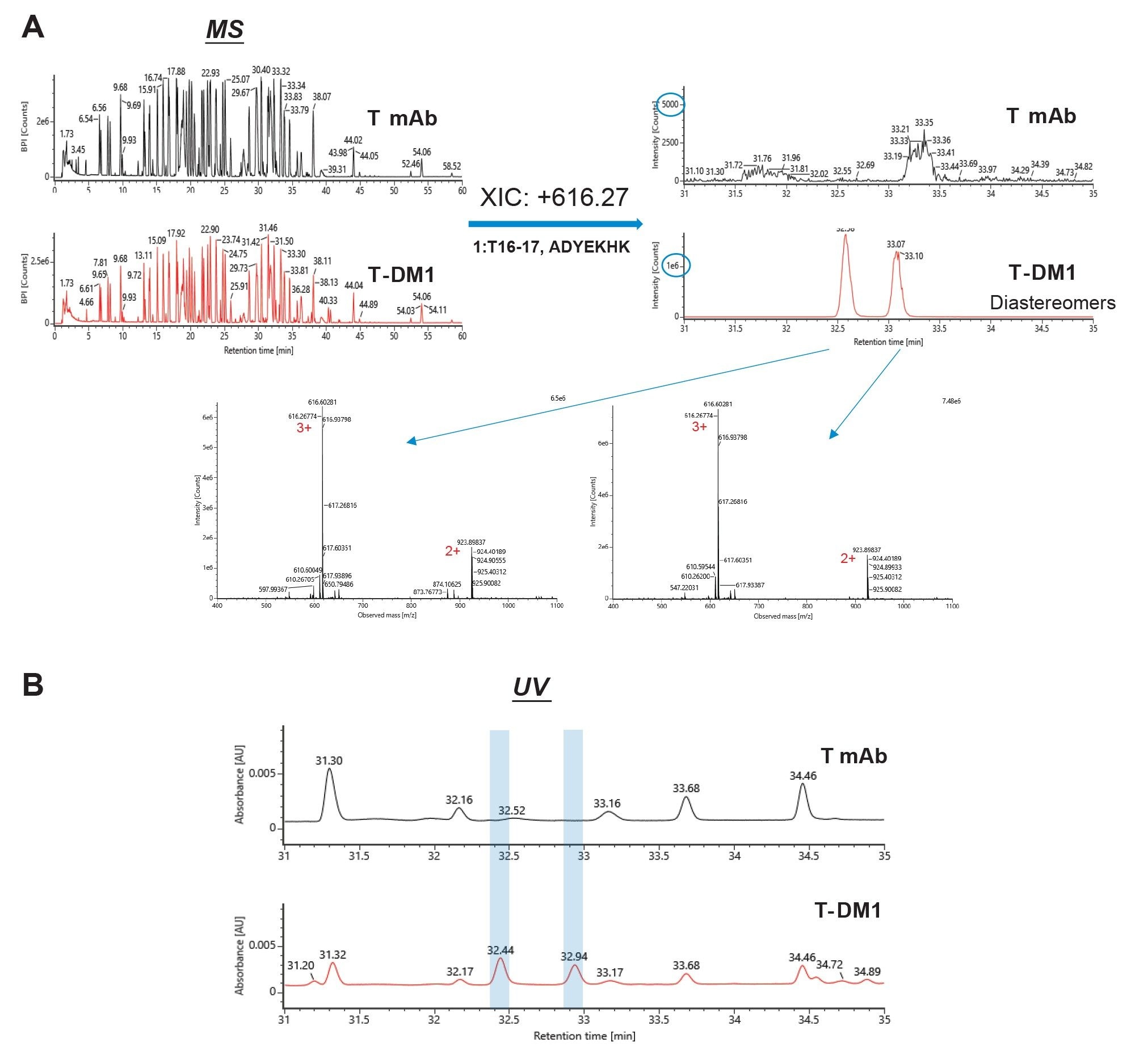
Figure 3 shows MS/MS spectra of the drug-conjugated light chain T16–17 fragment ions. As shown in the Fragmentation View of waters-connect software, y1-ion, y2-ion, b2-ion, and b3-ion are confirmed, strongly suggesting that the drug-conjugation site is the 5th amino acid (lysine) in the peptide sequence. Signature fragment ions specific to drug-conjugated peptide are also present (m/z in orange box), and the reputed structures are shown on top of the spectra. 4,5
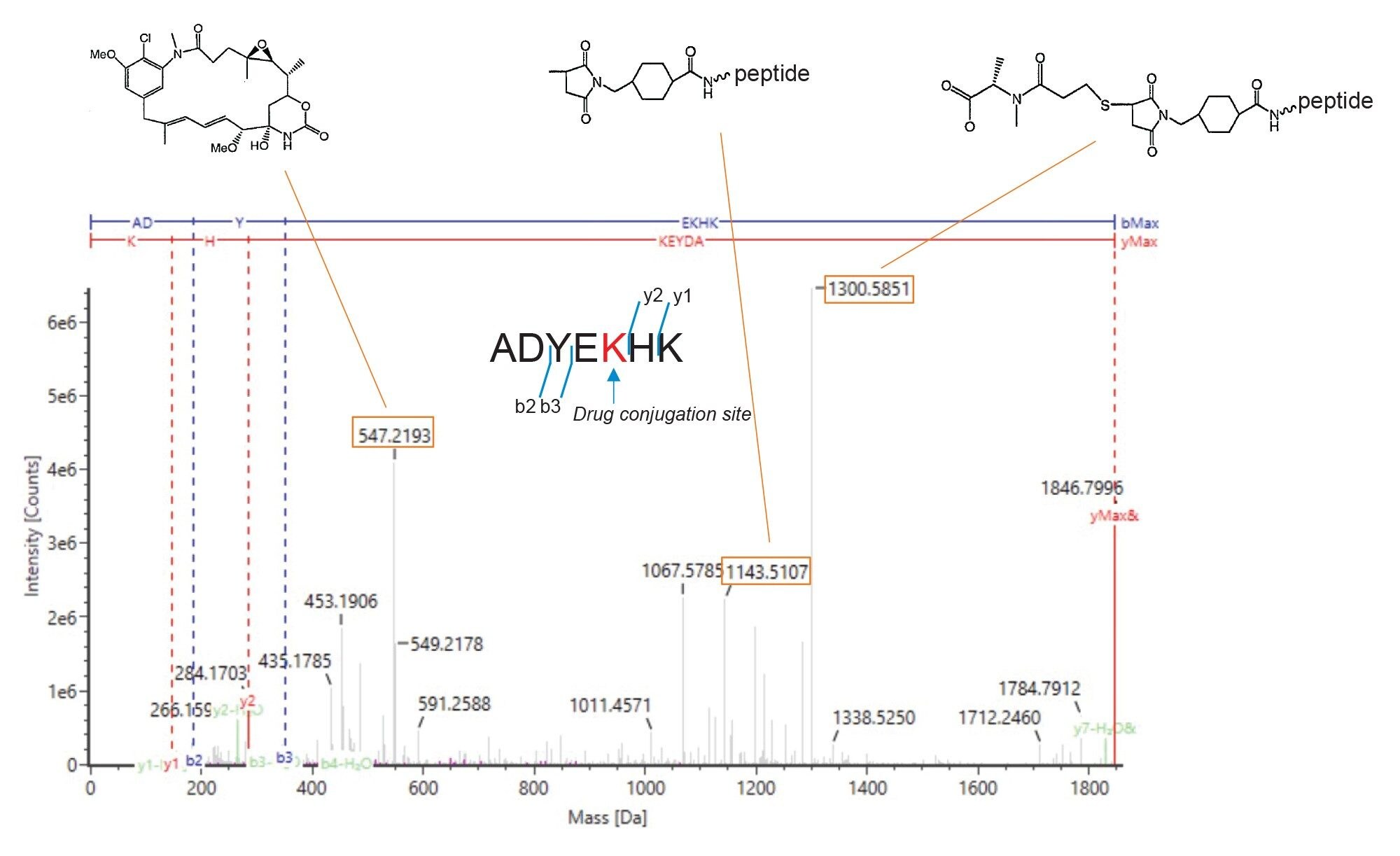
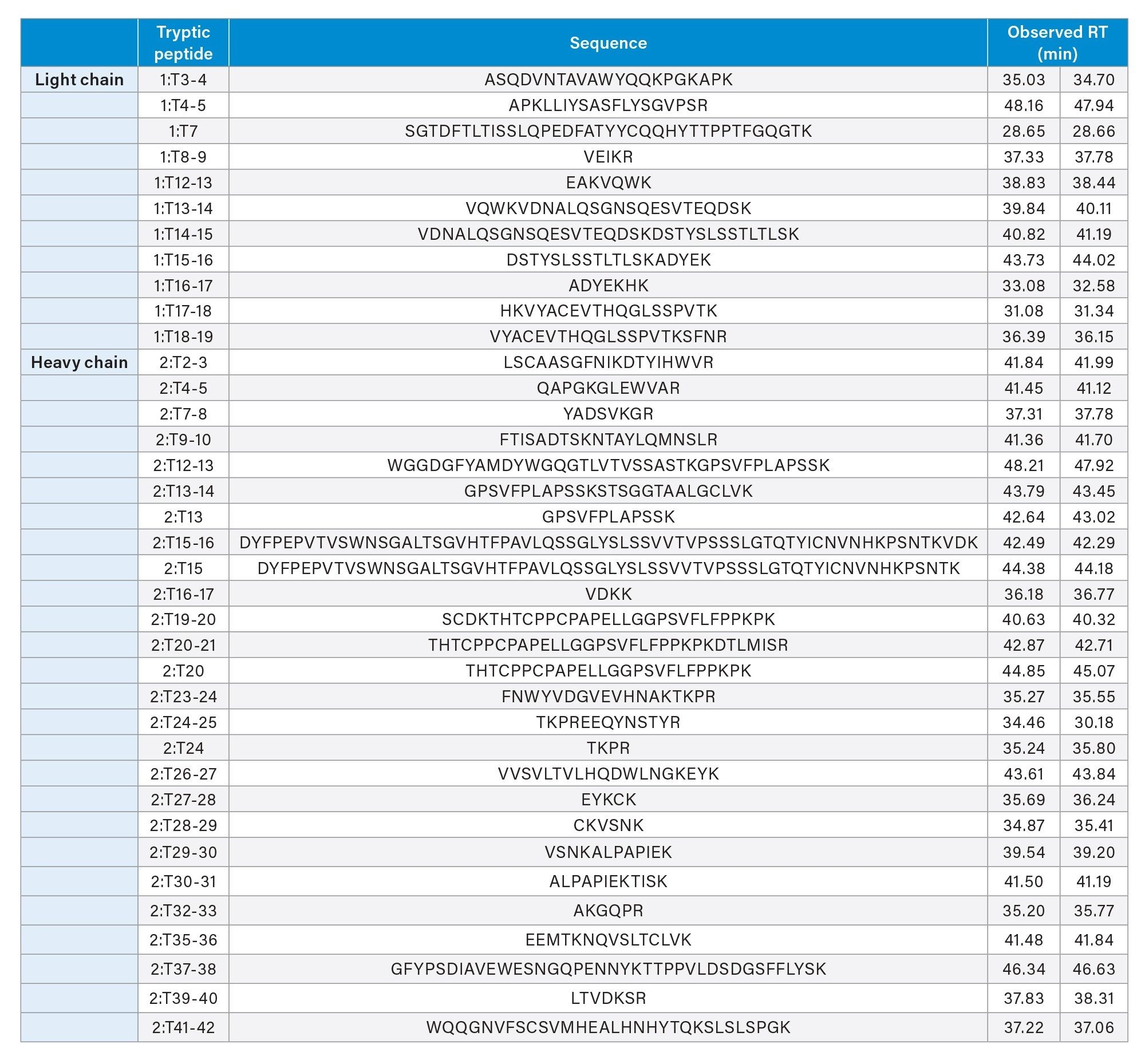
Table 1 shows a list of drug-conjugated peptides and their sequences. Most of the lysines are conjugated, consistent with previous results.4
It is worth noting that the coverage for T mAb and T-DM1 peptide map is 92% and 95%, respectively, with the predicted trypsin cleavage sites. When the drug conjugation site is taken into consideration, the coverage increases to 98% for T-DM1. This shows that PeptideWorks Tryptic Protein Digestion Kit can be a useful tool for mAb and ADC peptide mapping work.
Results and Discussion
Conclusion
PeptideWorks Tryptic Protein Digestion Kits can be used for ADC peptide mapping with automatable sample preparation procedure under 2.5 hours. Compared to the peptide map of the mAb without drug conjugation, additional peaks are observed on the ADC sample in the more retained region of the chromatogram. Using MS data, some diastereomer peptide pairs, resulted from two possible stereochemical configurations during conjugation reaction, are confirmed on the ADC sample.
References
- Hanna C.M., Danaceau J.P., Koza S.M., Shiner S., Trudeau M. Quick and Robust Sample Preparation for Tryptic Peptide Mapping With the PeptideWorks™ Kit Using Simple, Automatable Workflows. Waters Application Note. 720008019. July, 2023.
- PeptideWorks Tryptic Protein Digestion Kit Care and Use Manual. 720007980. October, 2024.
- RapiZyme Trypsin Care and Use Manual. 720007851. February, 2023.
- Chen L., Wang L., Shion H., Yu C., Yu Y.Q., Zhu L., Li M., Chen W., and Gao K. In-depth structural characterization of Kadcyla® (ado-trastuzumab emtansine) and its biosimilar candidate. MABS, VOL. 8, NO. 7, 1210–1223, October, 2016.
- Luo Q., Chung H. H., Borths C., Janson M., Wen J., Joubert M. K., and Wypych J. Structural Characterization of a Monoclonal Antibody−Maytansinoid Immunoconjugate. Anal. Chem. 88, 695–702, January, 2016.
720008664, January 2025