For research use only. Not for use in diagnostic procedures.
This is an Application Brief and does not contain a detailed Experimental section.
This application brief demonstrates the applicability of 1 mm scale chromatography for proteomic analyses, exhibiting a six-fold increase in throughput while maintaining similar identification results of that achieved with nanoscale chromatography.
One millimeter (1 mm) scale chromatography coupled to the SYNAPT XS using HDMSE analysis for complex proteomic sample analysis.
Today, there is an increased interest in clinical proteomics and its application for the analysis of large population cohorts. Traditional discovery proteomics typically uses nanoflow chromatography with 75 μm internal diameter columns using a flow rate between 0.2 and 0.5 μL/min, providing the high sensitivity and coverage for complex proteomic digests. However, long gradients and equilibration times are disadvantageous. For one sample, the total run time is typically between 60 and 180 min. Although the design of the LC has been largely improved for usability, for instance with finger tight fittings (i.e., Waters ZenFit Connectors), nanoscale chromatography remains challenging to use and troubleshoot, restricting its applicability for large cohort analysis. Recently, with improved mass spectrometry resolution and sensitivity, microflow scale chromatography using 300 μm column internal diameters with flow rates between 7 and 12 μL/min have been implemented to analyze large cohorts of digested human plasma samples.1,2,3 Here, we show the utility of 1 mm scale chromatography coupled to the SYNAPT XS Mass Spectrometer to analyze mid-complexity samples such as E. coli and human plasma digests in a reduced timeframe (i.e., 15-min gradient), while maintaining comparable protein/peptide identifications.
Two sample types were used to evaluate the 1 mm scale chromatography performance: human plasma and a Waters digested E. coli cytosolic extract (p/n: 186003196).
The human plasma samples corresponding to seven conditions and one pool control (QC) were reduced, alkylated, tryptically digested, and concentrated using solid phase extraction (SPE) following the Waters ProteinWorks protocol (p/n: 176003688). iRT peptides (Biognosys) were used as an internal reference with 1 μL spiked into 50 μL of sample. The E. coli digest were prepared at a concentration of 1 μg/μL and 100 ng/μl.
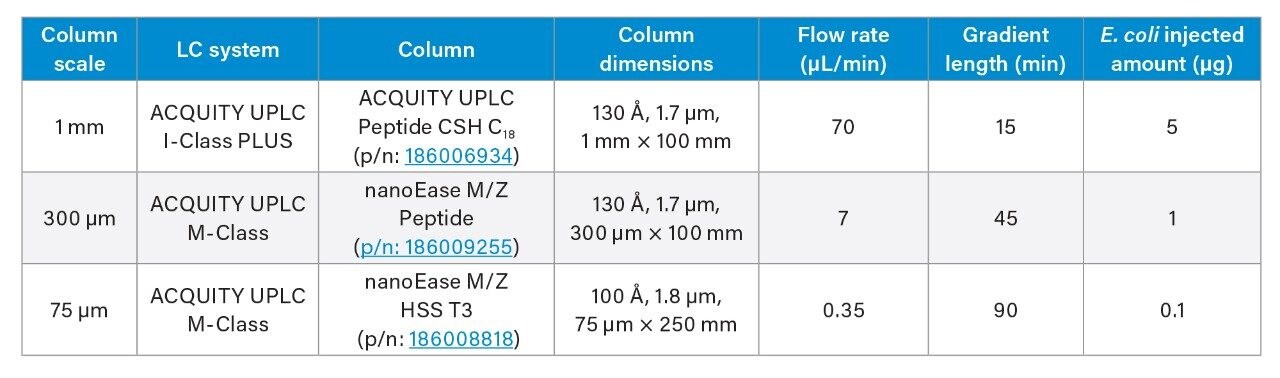
The different scales of chromatography used are described in Table 1. The mobile phases consisted of 0.1% formic acid in water (mobile phase A) and 0.1% formic acid in acetonitrile (mobile phase B). Peptides were eluted using a gradient from 3–35% mobile phase B; 3.5 μg of plasma were injected in triplicate (1 mm setup only). The quantity of E. coli injected for the various column geometries is outlined in Table 1.
Data were acquired on the SYNAPT XS Mass Spectrometer operating in HDMSE (positive ion mode) and processed using Progenesis QI for Proteomics v4.2 and Skyline v20.1 (MacCoss Lab, University of Washington, USA).
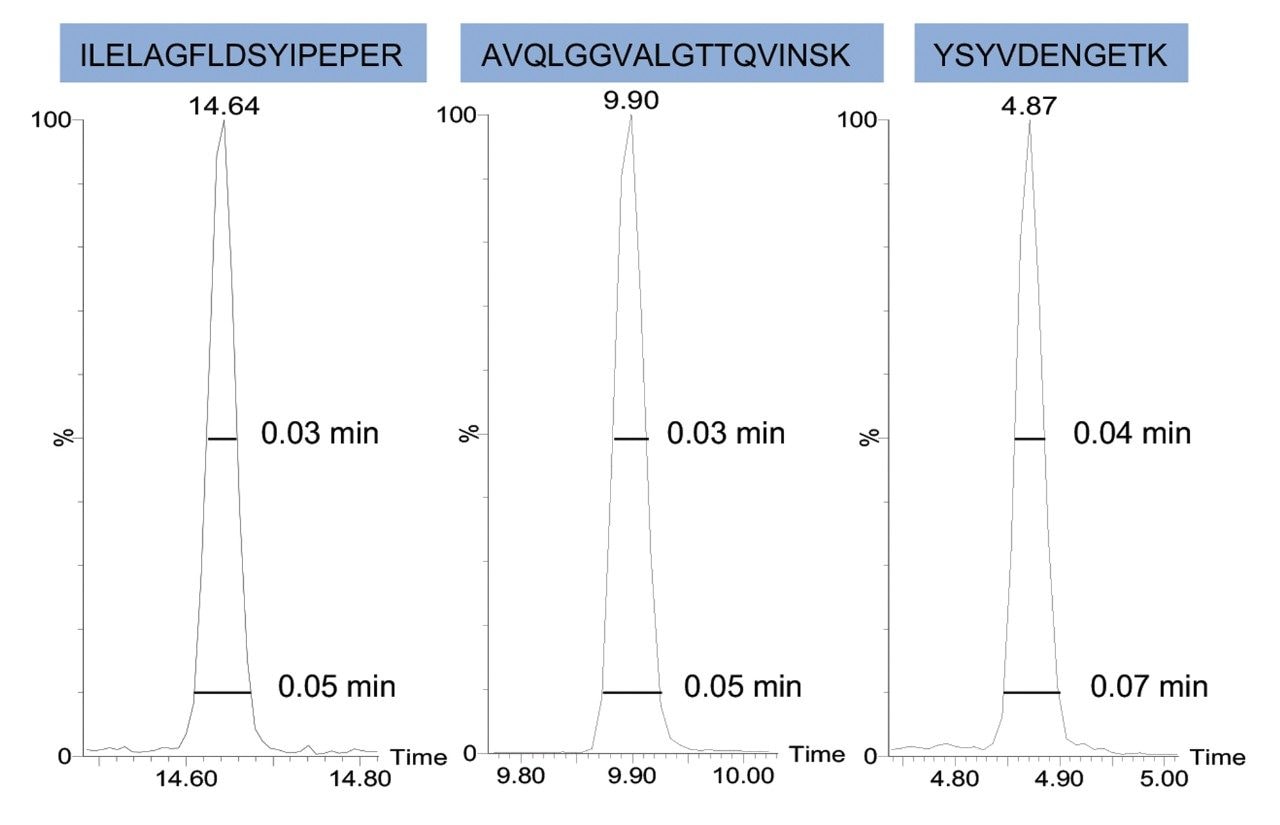
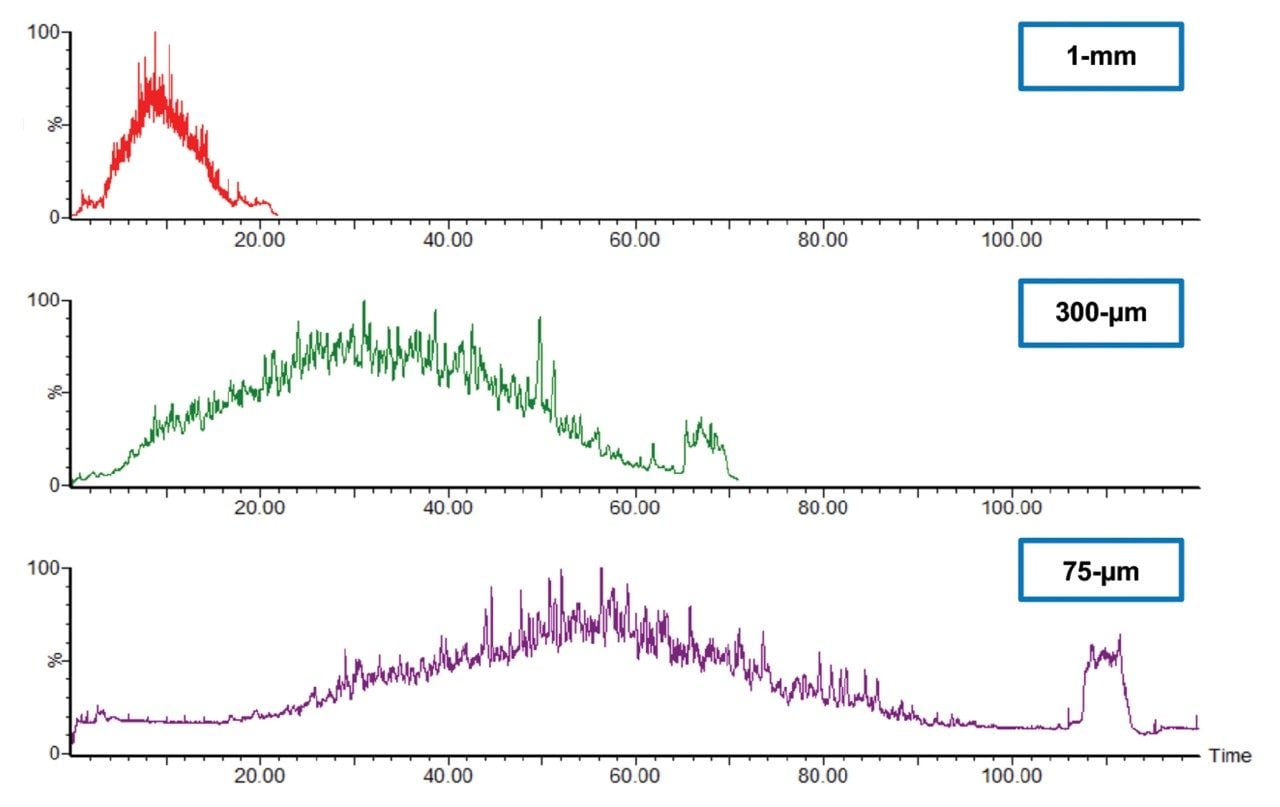
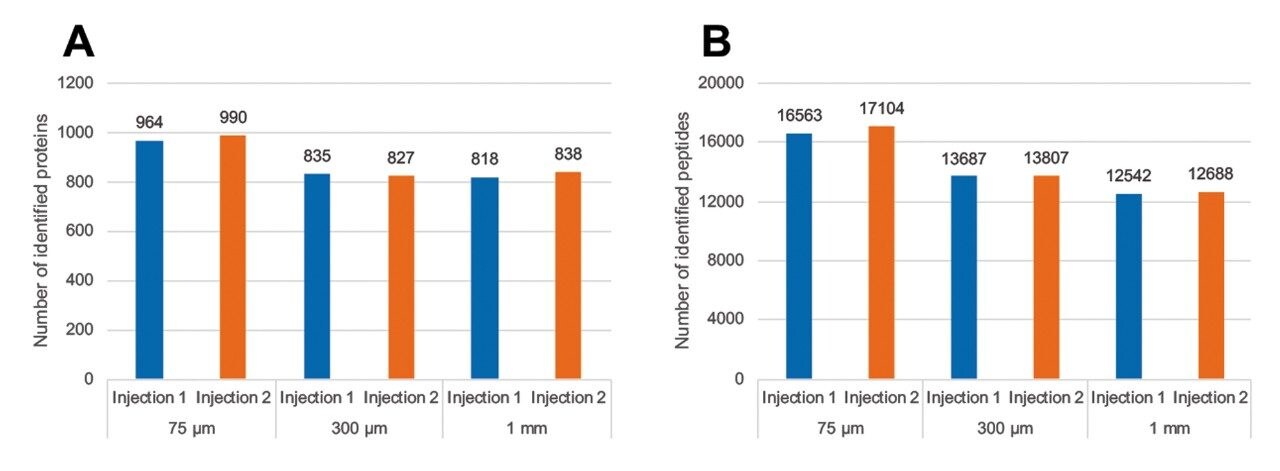
Three E. coli peptides were extracted at different retention times across the 15-min gradient to show the high chromatographic resolution obtained from 1 mm scale chromatography. Peak widths at full width half-maximum (FWHM) and 10% peak heights were shown to be 0.03–0.04 min and 0.05–0.07 min respectively (Figure 1). Representative chromatograms for each column I.D. are provided in Figure 2. The number of E.coli proteins and peptides identified for each of the columns evaluated are shown in Figure 3. Interestingly, the difference in protein identifications were shown to be minimal with increasing column I.D., while showing increased benefits in terms of throughput, whereby the gradient was reduced by six times from 90 min (75 μm) to 15 min (1 mm).
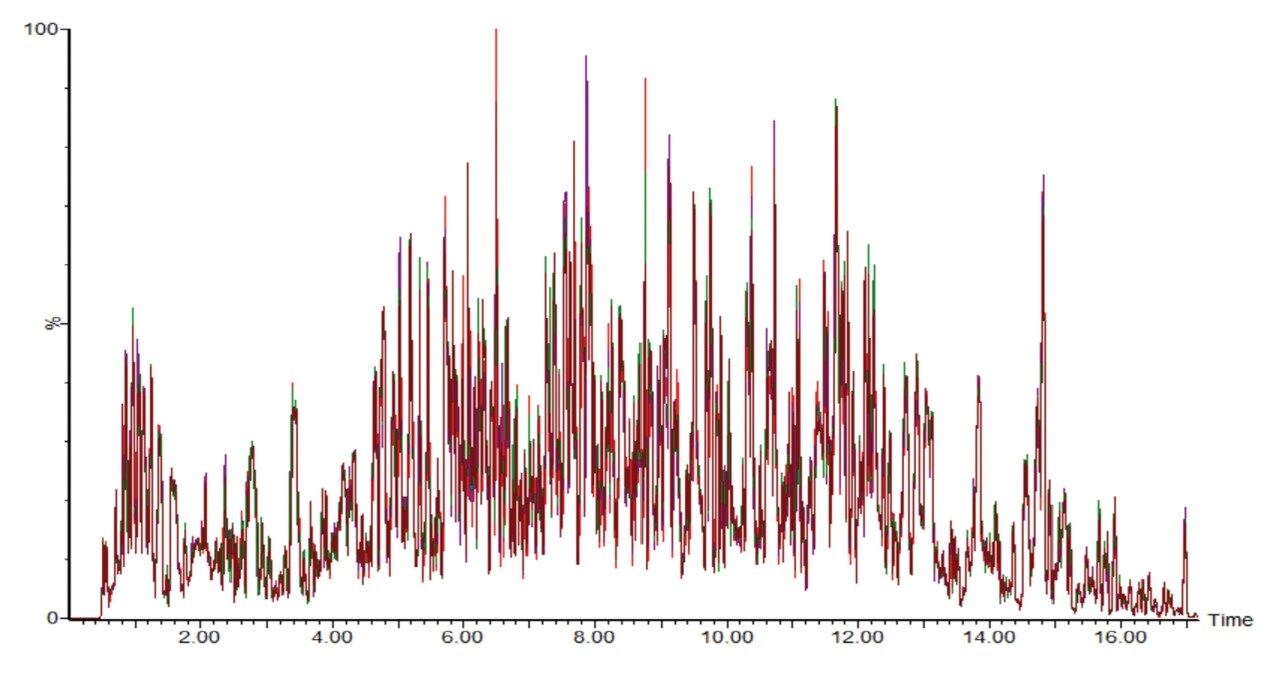
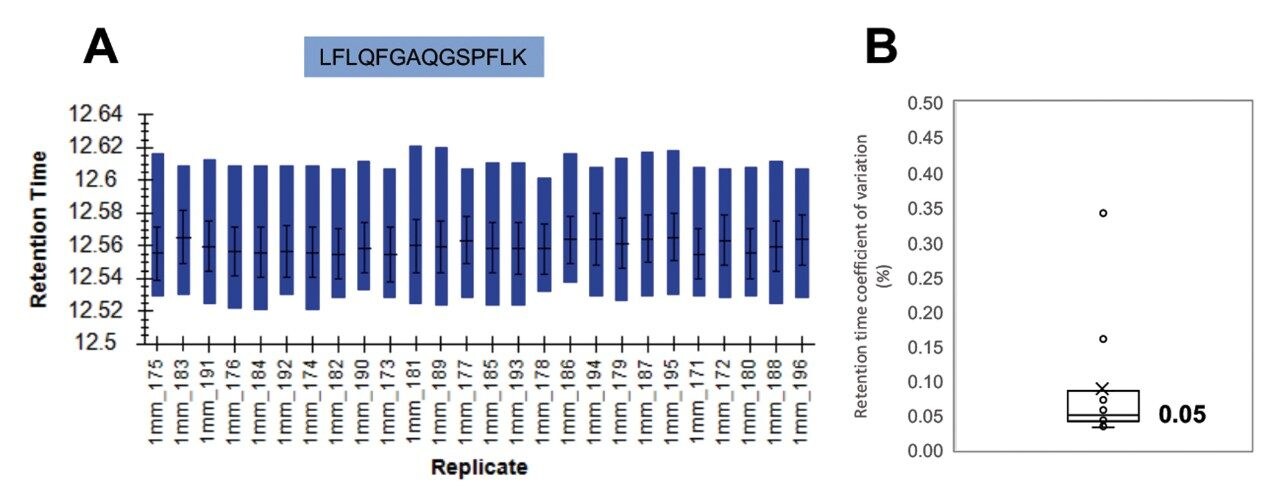
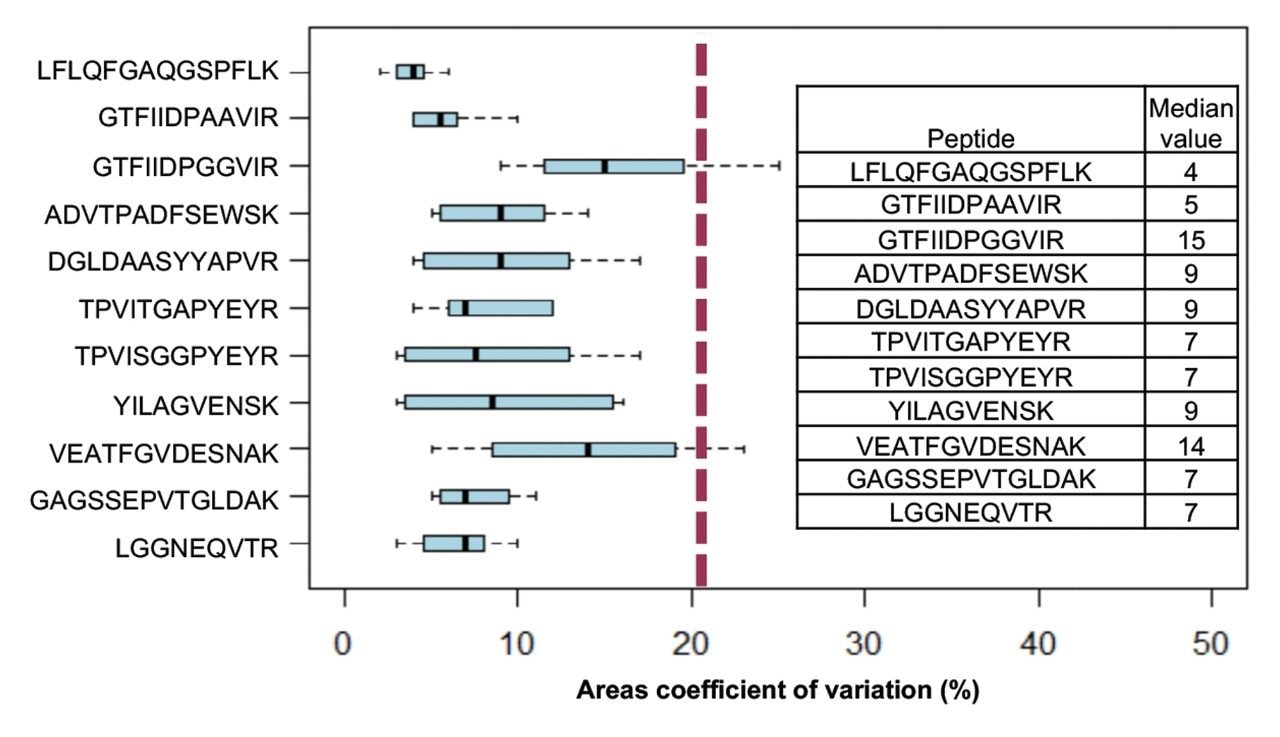
Excellent chromatographic reproducibility based on 1 mm scale was obtained for the plasma samples and is shown with the overlay of five pooled QC samples (Figure 4). The median coefficient of variation (CV) in retention time for the 11 iRT peptides across all samples was observed to be 0.05% (Figure 5). Moreover, the peak area CV for each iRT peptide in each sample type is shown to be less than 20% for the majority of peptides (Figure 6). Altogether, 533 plasma related proteins were identified with a false discovery rate (FDR) <1%.
Utilizing 1 mm scale chromatography coupled to the SYNAPT XS Mass Spectrometer has shown to provide high quality data for moderately complex proteomic analyses (i.e., E. coli and plasma digests). The number of protein and peptide identifications is comparable with nanoflow configurations. The acquisition time is drastically reduced from 90 min (75 μm) to 15 min (1 mm) allowing for an approximate six-fold increase in sample throughput, making it suitable for large cohort studies. Moreover, the set-up has been shown to be highly reproducible in terms of retention times and peak areas.
720006833, April 2020