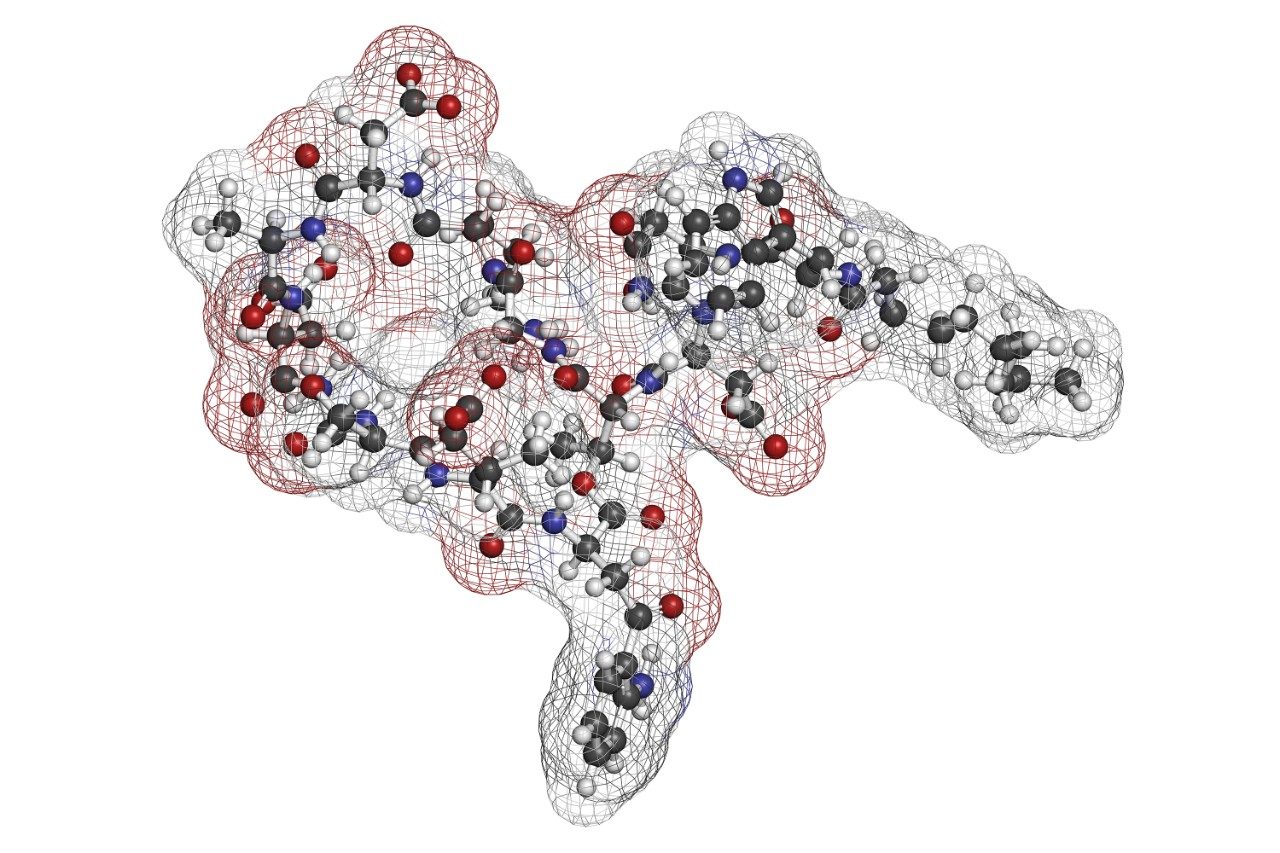
In this application note, we describe a solution that uses an information-rich data acquisition mode, which effectively deals with the complexity of modified peptides. This enables key metabolic data to be generated unambiguously while tackling complex peptide studies. The use of DIA combined with ion mobility (HDMSE) improves upon the selectivity provided by MS-only techniques. The addition of routine Collision Cross Section (CCS) measurements makes HDMSE a powerful identification acquisition mode, as well as an ideal separation technique.
The incorporation of unnatural amino acids, backbone modifications, conjugations, and cyclizations is a commonly used strategy to improve the efficacy and ADME profile of peptide therapeutics. Analyzing these complex peptides (often natural products which may be synthetically modified) requires overcoming a variety of analytical and technical challenges, including challenging chromatography, non-specific binding, limited or complex fragmentation, and the prediction and review of many catabolites and/or novel metabolites. To ensure safe and efficacious therapies, the metabolic fate of potential drug candidates needs to be thoroughly and rapidly investigated using LC-MS coupled with informatics capable of identifying and characterizing their profiles.
Current approaches for analyzing complex modified peptides suffer from challenges that include:
Here, we describe a solution that uses an information-rich data acquisition mode, which effectively deals with the complexity of modified peptides. This enables key metabolic data to be generated unambiguously while tackling complex peptide studies. The use of DIA combined with ion mobility (HDMSE) improves upon the selectivity provided by MS-only techniques. The addition of routine Collision Cross Section (CCS) measurements makes HDMSE a powerful identification acquisition mode, as well as an ideal separation technique. For those projects requiring definitive product ion scans, we describe the use of simultaneous product ion confirmation scans (PICS, IMS MS/MS) for metabolites with complex product ion spectra to enable the highest confidence in elucidating structures. Loss of metabolites and signal response can be mitigated through the use of low binding protocols or products such as QuanRecovery vials.
All data for daptomycin, dalbavancin, and lanreotide were screened for metabolism using an ion mobility-enabled HDMSE acquisition workflow and processed using Mass-MetaSite (MMS) and WebMetabase (WMB) macromolecule software packages (Lead Molecular Design, S.L, Sant Cugat del Vallés, Spain). HDMSE and HDMSE + product ion data were reviewed using the UNIFI screening application.
Samples were incubated and processed according to methodology reported by Radchenko et al.1 Briefly, chymotrypsin (1 mg/mL) (MilliporeSigma, St.Louis, MO) was dissolved in simulated intestinal fluid (SIF) containing pancreatin (Ricca Chemical, Arlington, TX). 50 μL incubations were prepared in 300 μL QuanRecovery vials (Waters Corporation, Milford, MA). One mM stock solutions of daptomycin, dalbavancin or lanreotide (MilleporeSigma, St.Louis, MO) were prepared. 0, 5, 15, 45, 120, and 300 min timepoints for daptomycin and 0 and 300 min for dalbavancin and lanreotide were prepared at a starting concentration of 10 μM. Each timepoint was quenched with two volumes cold acetonitrile containing 1% ammonium hydroxide (by volume).
|
LC conditions |
|
|---|---|
|
LC system: |
ACQUITY UPLC I-Class PLUS with FTN |
|
Vials: |
QuanRecovery with MaxPeak |
|
Column: |
ACQUITY UPLC Peptide BEH C18 300 Å, 1.7 μm |
|
Column temp.: |
60 °C |
|
Sample temp.: |
8 °C |
|
Injection volume: |
2 μL |
|
Flow rate: |
0.5 mL/min |
|
Mobile phase A: |
Water, 0.1% formic acid |
|
Mobile phase B: |
Acetonitrile, 0.1% formic acid |
|
Gradient: |
2 to 60% B over 7 min; 60 to 90% B from 7 to 7.5 min; 90% B hold from 7.5 to 8.5 min; and re-equilibrate at 2% B from 8 to 10 min. |
|
MS system: |
Vion IMS QTof Mass Spectrometer |
|
Ionization mode: |
ESI+ |
|
Acquisition range: |
50–2000 m/z |
|
Capillary voltage |
1.5 kV |
|
HDMSE collision energy: |
4 (low energy) 15–70 eV ramp (high energy) |
|
HS-MSMS collision energy: |
12–40 eV ramp @ 50 m/z to 22–80 @ 2000 m/z |
|
Cone voltage: |
80 V |
UNIFI Scientific Information System, version 1.9 SR4 API enabled Mass-MetaSite 3.4.9, WebMetabase 4.03 (Lead Molecular Design, S.L, Sant Cugat del Vallés, Spain)
All data were acquired in UNIFI and processed using the Mass-MetaSite batch processor. For each peptide, time points with blanks and standards were processed automatically (in the US) and uploaded to a WebMetabase server (based in the UK), and reviewed via a web browser interface.
The primary goal of this study was to verify that data for complex peptide structures could be rapidly acquired using HDMSE acquisition on a Vion IMS QTof MS and subsequently processed for catabolism identification using Mass-MetaSite and WebMetabase directly using the UNIFI Application Programming Interface (API).2
A cross section of peptides were tested containing various non-natural substitutions, adducts, and increasingly complex backbones and cyclizations within the macromolecular structures (Figure 1). For all peptides, the top three metabolites were reported and for daptomycin, a larger discussion of metabolites and CCS values generated is provided. Table 1 shows the complexity observed in the peptides tested and summarizes the structural modifications that can be handled by Mass-MetaSite and WebMetabase. Simpler peptides shown in Table 1, such as GLP-1 and somatostatin, are not discussed here, but have been previously published by several labs.3,4
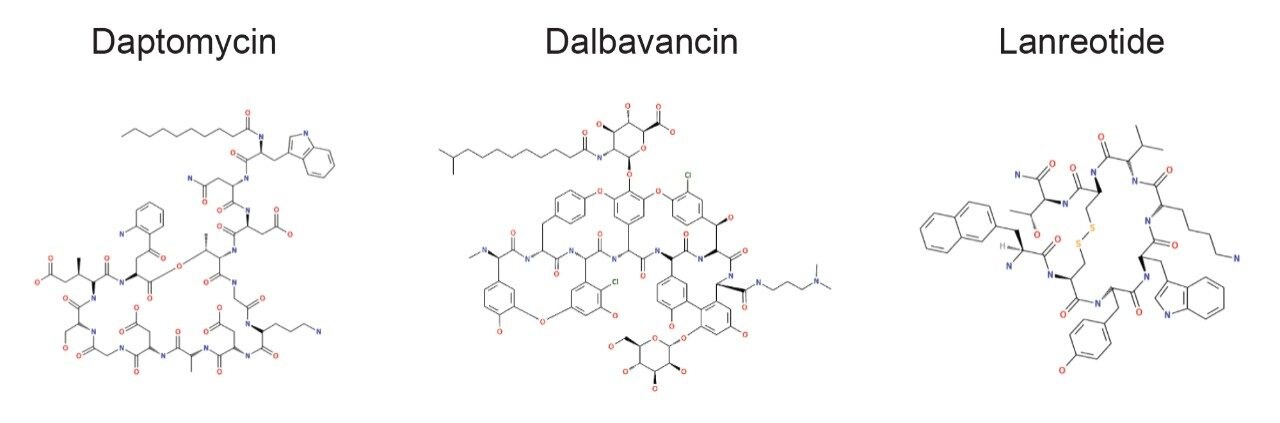
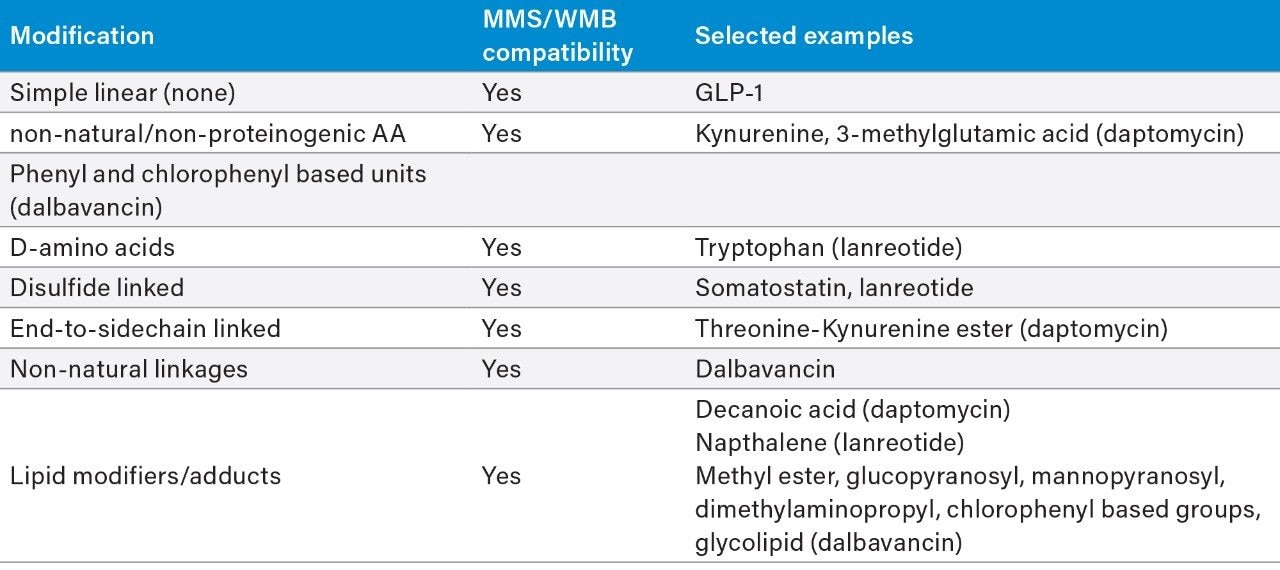
A small to moderate amount of catabolism was detected for all peptides. Daptomycin showed higher turnover after 300 min (>60%), while other peptides were found to be more stable and showed <10% turnover. The top three catabolites were reported for each peptide (Table 2). Daptomycin was chosen as a model compound to further demonstrate capabilities in DIA/DDA acquisition modes and software data processing.
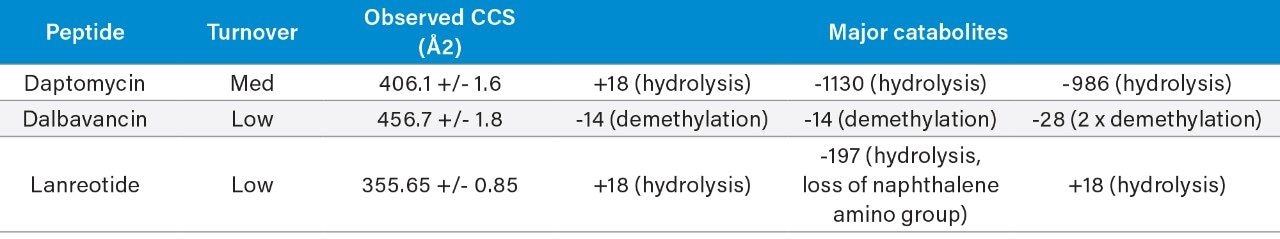
A summary of the major catabolites for daptomycin is reported in Figure 2, along with the summary trace at 300 min. Data were reviewed with filters applied that excluded singleton metabolites (those present only at one timepoint) and results that were > +/- 2 mDa different from expected values. All other metabolites were reviewed for probable matches. Cluster plots (time courses) were reviewed to ensure profiles were consistent with turnover. Data were reviewed for plausible CCS values that corresponded to a given structure change. For example, the loss of an amino acid or a fatty acid tail was expected to result in a slightly smaller CCS at a given charge state, while ring opening was generally expected to decrease mobility and increase CCS. Consistency of mass accuracy, retention time, and CCS accuracy were also tracked by the software and used to help confirm metabolic profiles and further remove matrix false positives.
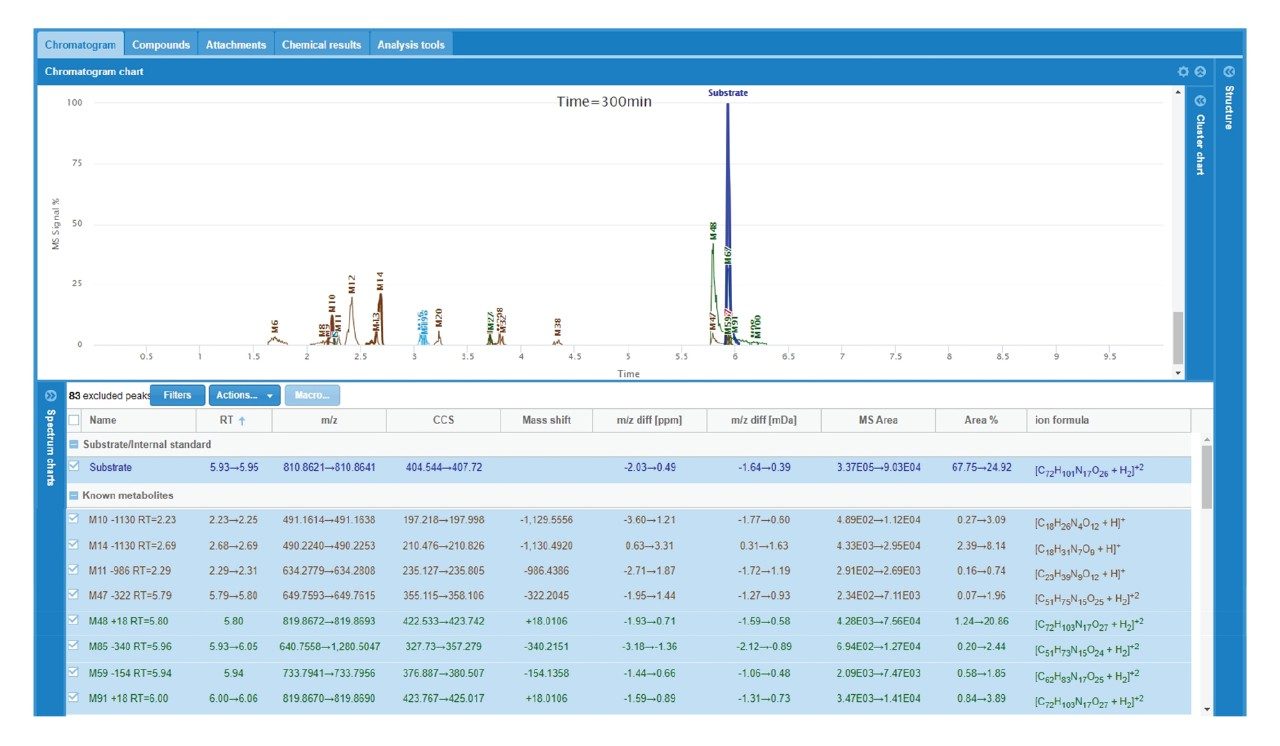
The increase and/or disappearance of major species are displayed in the software as a cluster plot. An example of response versus time of major species for daptomycin is shown in Figure 3. WebMetabase automatically filters the data to show only metabolites that appear in more than one sample through the remove singletons option, further refining table results.
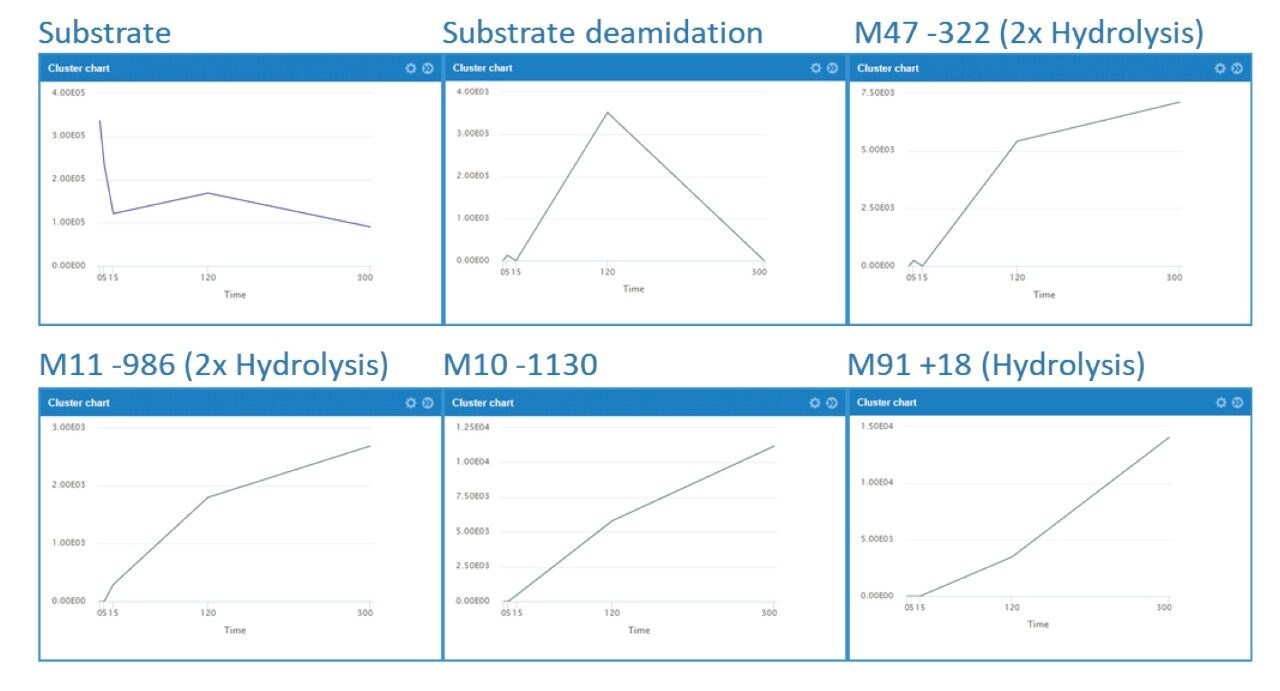
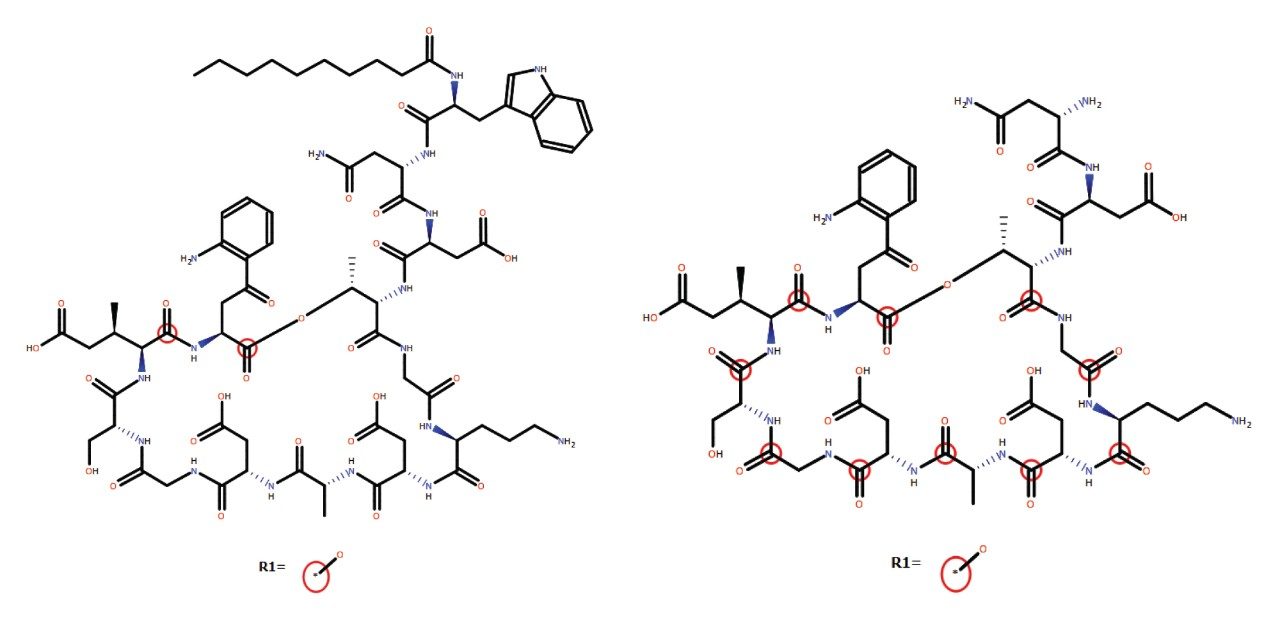
Several peaks corresponding to amide hydrolysis were observed and putative assignments shown in Figure 4. For the major hydrolysis peak, M91, the site of biotransformation was localized to a cyclic cleavage product involving the hydrolysis of the ester, or a close by amide bond between the 3-anthraniloylalanine (kynurenine) and the 3-methylglutamic acid residues. Both sites have been previously reported as primary sites of metabolism.5 Cleavage of the terminal fatty acid tail (M59-154) in isolation and in combination with the terminal tryptophan (M47-322) were also observed and reported by the software in these samples.
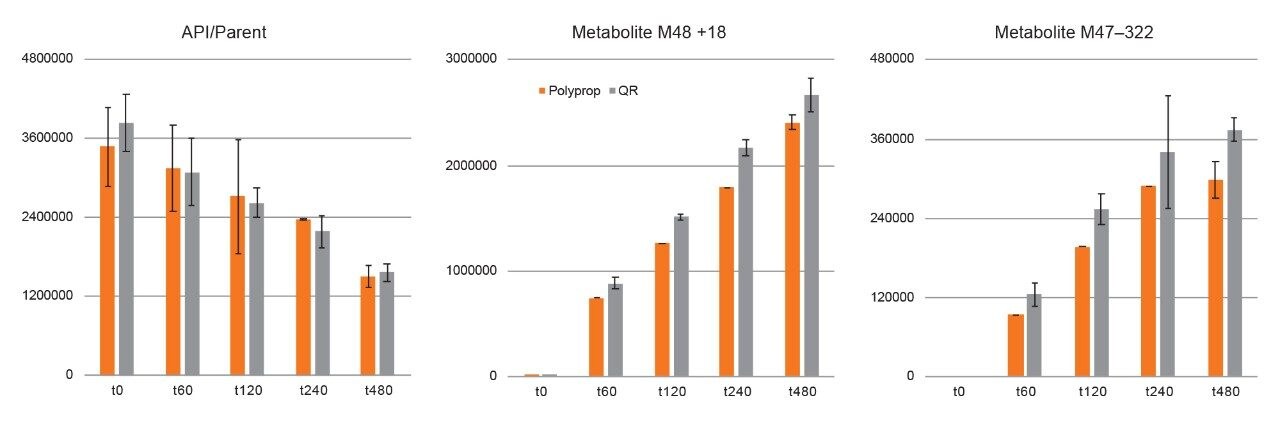
Experiments using QuanRecovery vials showed equivalent performance or better recovery for the API and metabolites. For this series of experiments, the incubation time was extended to 480 minutes. API recovery was found to be equivalent for polypropylene and QuanRecovery vials. For several metabolites such as M48 and M47, the recovery was consistently better using QuanRecovery vials with an average of 18% and 26% higher response, respectively.
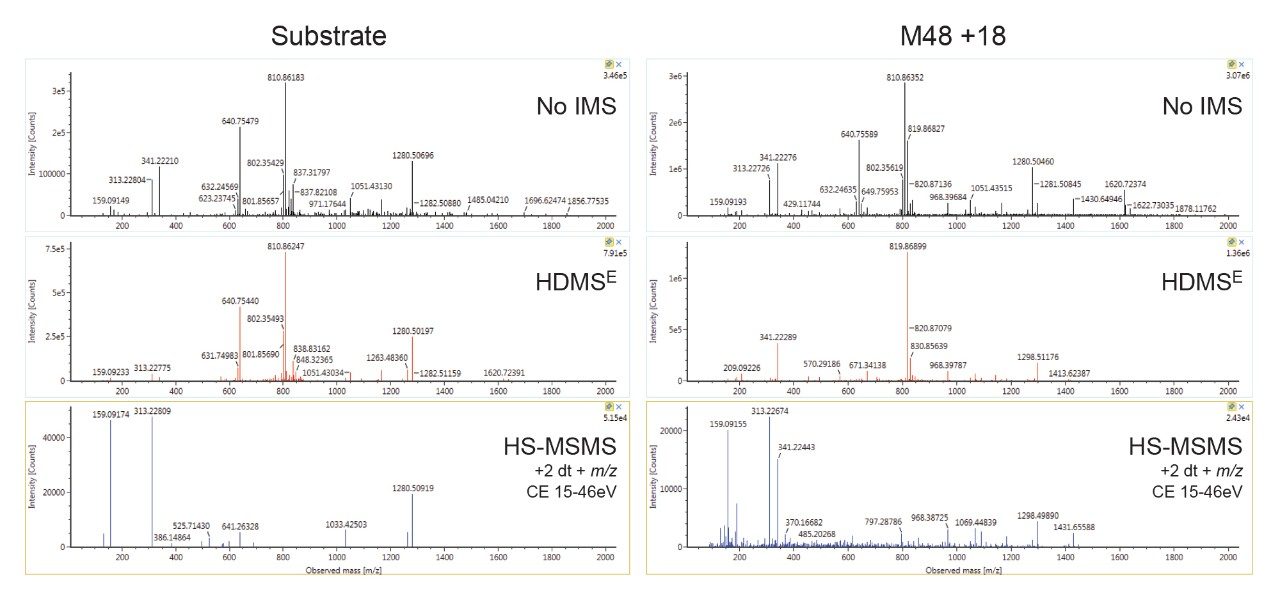
The addition of directed scanning functions to DIA methods enables full confirmation for ions of interest. Product ion confirmation scans (PICS) can be enabled for MSE or HDMSE analyses and spectra may be viewed in UNIFI. Figure 6 shows representative examples of data acquired without IMS processing, with IMS processing (HDMSE) and with HS-MSMS product ion spectra (ion mobility precursor selected MSMS) enabled. The data shows that IMS significantly enhances the spectrum by improving specificity and removing ions present in the unfiltered top trace, cleaning up noisy baselines and improving the quality of spectra. The use of drift time alignment can be used to combine multiple charge states into a single rich spectrum, containing all fragments produced by all major charge states. HS-MSMS data was optimized further (higher CE, mass dependent ramp) to generate a high-quality spectrum for the major (+2) charge state peaks. This results in a further refined spectrum with better peak quality, particularly in the mid region of the spectrum, which is more straightforward to interpret.
Metabolites for 3 FDA-approved cyclic peptide drugs of varying structural complexity were identified and tracked using the WebMetabase macromolecule workflow. Ion mobility afforded additional resolution that was shown to help further discriminate, characterize, resolve, and track metabolites from interfering matrix ions having similar m/z but significantly different ion mobility. Ion mobility benefits both DIA and DDA approaches for generation of high-quality spectra. CCS enabled HDMSE provided tools for tracking metabolites across multiple samples and instruments. Product ion confirmation scans (PICS) offer an easy way to build on the strength of the HDMSE approach by providing a capability for adding true ion mobility and mass directed HS-MS/MS scans to DIA analysis.
The use of an ion mobility capable software package to process and review RT, m/z, MS-MS, and CCS information, along with cluster plots and integrated structural ID tools, help users quickly decide if peaks are real and whether they warrant further investigation. Proper handling of peptide therapeutics is essential and the use of high-performance non-binding products such as QuanRecovery vials enables better and more consistent recovery of detected metabolites.
Combining ion-mobility enabled HDMSE data with a powerful software solution tailored for biotherapeutics enabled comprehensive metabolite identification of complex cyclic peptides needed for the development of efficacious biotherapeutics.
HDMSE – MSE is a full scan acquisition mode that uses alternating collision energies and aligns the precursor and fragment ion information for all species.6 The low energy channel, is optimized for normal full scan transmission; and the high energy channel is optimized for precursor transmission. High Definition (HD) modes adds TWIMS (Travelling Wave Ion Mobility-based separation to both functions. Drift time information is captured by recording the drift time of all ions that pass through the ion mobility separation device and improves DIA data quality.
HS-MS/MS or HS-DDA MSMS – Once the drift time is known (determined from a routine HDMS scan mode), targeted MS/MS scans can select precursors exiting the ion mobility cell at a given time. HS (High Selectivity) scans further increase the specificity of precursor ions passed through for MS/MS using ion mobility. This type of scan is enabled when PICS is selected for an HDMSE analysis.7
CCS – Collision Cross Section – Represents the average cross section area for a molecule as it passes through a gas cell. Larger, extended structures and higher charge state species take longer to pass through a gas cell versus smaller more compact structures for the same given m/z creating an additional mechanism for ion separation.8 For CCS on a Vion IMS QTof, drift times are calibrated against a known set of standards (using Major Mix, PN 186008113) during routine calibration. CCS values are calculated automatically for all analyses run using HD modes in UNIFI.
720006850, April 2020