For research use only. Not for use in diagnostic procedures.
This study highlights the qualitative and quantitative advantages of SONAR for high throughput proteomics.
Quantitative proteomics often incorporates the use of stable labeled isotopes (SILs) in order to provide absolute quantification. Recent advancements have seen the introduction of peptide panels allowing the quantification of more than 500 proteins in plasma sample sets. However, this is technically challenging when attempting to acquire the data using more traditional MS acquisition modes such as multiple reaction monitoring (MRM), since the duty cycle of the instrument is compromised and therefore results in under sampling of the data. An alternative approach is to apply SONAR, a data independent analysis (DIA) methodology, allowing for high throughput while also ensuring high specificity and maintaining quantitative performance. SONAR has previously been described,1,2 highlighting the utilization of a fast scanning quadrupole, enabling the technique to be compatible with fast chromatography, high throughput workflows. Clinical research using proteomics is one high throughput example which requires the analytical benefits provided by SONAR. Here, we present the applicability of SONAR for high throughput, absolute quantification of plasma proteins from human samples consisting of controls, chronic obstructive pulmonary disease (COPD), and asthma.
Undepleted human plasma (Innovative Research, MI) originating from controls (n=6) and confirmed COPD (n=6) and asthma (n=6) source samples were reduced, alkylated, and tryptically digested overnight. Prior to LC-MS analysis, samples corresponding to their individual groups were pooled and spiked with PQ500 SIL peptides (Biognosys AG, Schlieren, Switzerland).
|
LC system: |
ACQUITY UPLC M-Class |
|
Column(s): |
Peptide CSH C18 130Å, 1.7 μm, 300 μm × 100 mm (p/n 186009255) |
|
Column temp.: |
55 °C |
|
Flow rate: |
7 μL/min |
|
Mobile phase A: |
Water (0.1% formic acid) |
|
Mobile phase B: |
Acetonitrile (0.1% formic acid) |
|
Gradient: |
3% to 40% B in 15 or 45 min |
|
Injection volume: |
5 μL (5 μg) |
|
MS system |
Xevo G2-XS QTof |
|
Ionization mode |
ESI (+) at 2.2 kV |
|
Cone voltage |
30 V |
|
Acquisition mode |
SONAR |
|
Acquisition range |
50 to 2000 m/z both functions (low and elevated energy) |
|
Acquisition rate |
0.5 s both functions (low and elevated energy) |
|
Quadrupole scan range: |
330 to 1100 m/z |
|
Isolation window |
20 Da |
|
Collision energy |
6 eV (low energy function) and from 14 eV to 36 eV (elevated energy function) |
|
Resolution |
30,000 FWHM |
LC-MS data were processed with Spectronaut Pulsar X (Biognosys AG, Schlieren, Switzerland). Data were searched against a human plasma specific spectral library, generated from SONAR processed data with Progenesis QI for Proteomics. Additional data visualization and statistical analysis was performed using Metaboanalyst.1
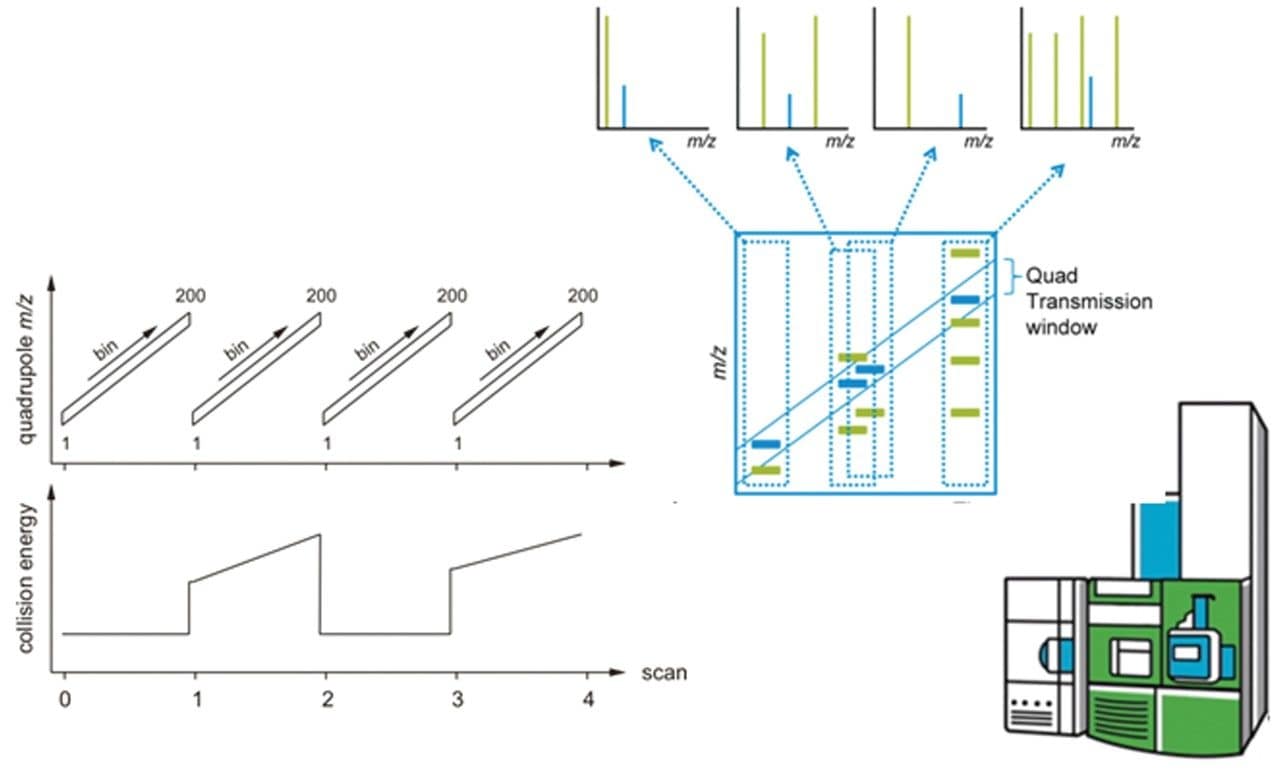
The principle of SONAR, a scanning quadrupole based DIA method, is illustrated in the left hand side image of Figure 1. In short, alternate datasets are acquired in low (MS1) and elevated (MS2) collision energy mode.2,3 During each low and elevated energy segment, the quadrupole isolation window is scanned linearly between two user-selected positions and 200 TOF spectra are acquired. The quadrupole scan duration is application/chromatographic peak width dependent and typically varies from 0.1 s to 1 s. In the elevated energy mode, the collision energy can be ramped between two values, which are selected to optimize fragmentation efficiency at each quadrupole position. The selectivity of the acquisition method is illustrated by the middle image where, dependent on the position of the quadrupole, i.e. transmission m/z window, precursor (even those close in mass) and product ions can be exclusively isolated.
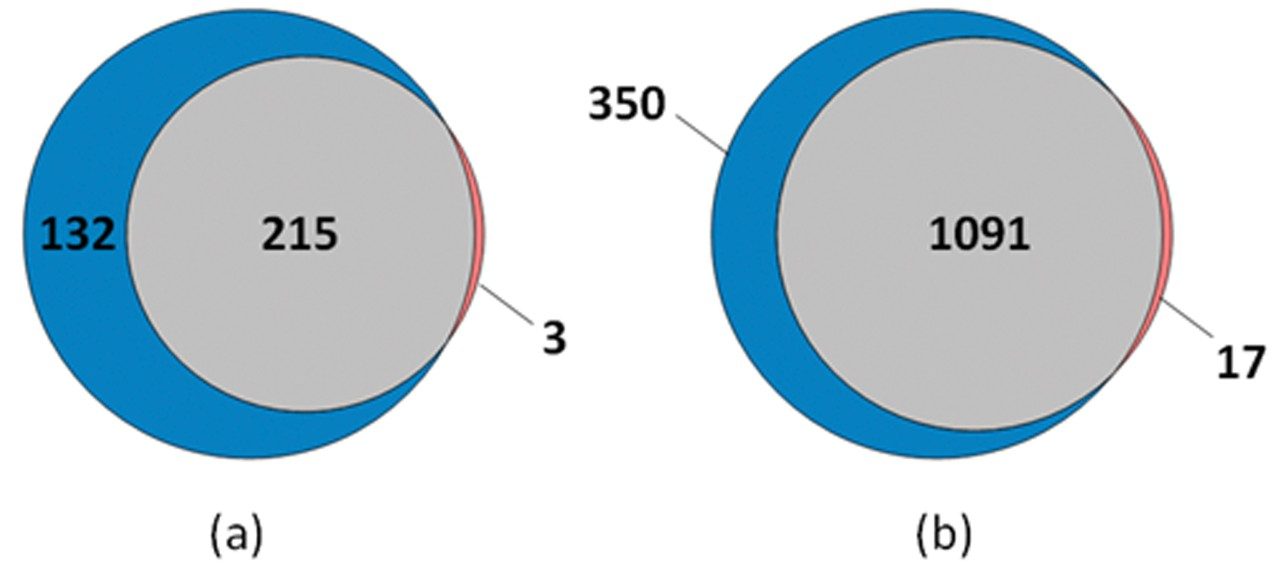
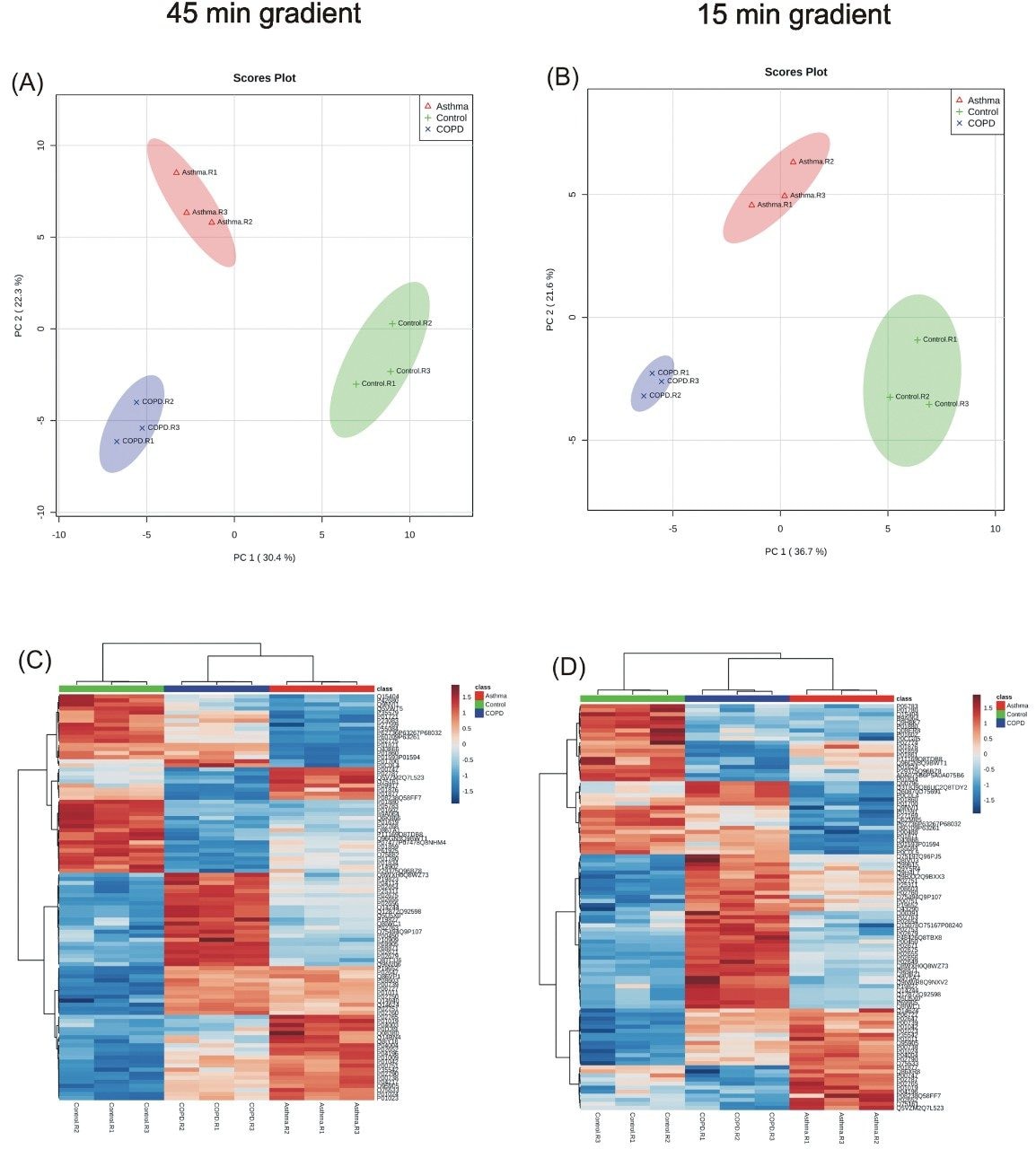
Data was collected for undepleted plasma which was separated over either 45 or 15 min gradients, resulting in 347 and 218 protein identifications respectively (Figure 2). Regardless of the gradient conditions employed, statistical analysis showed clear separation between the three cohorts, with all three technical replicates clustering together. Likewise, a comparison of protein abundances using hierarchical clustering on the basis of Euclidean distances, showed distinct clusters of proteins which produced similar levels of protein expression under both gradient conditions (Figure 3).
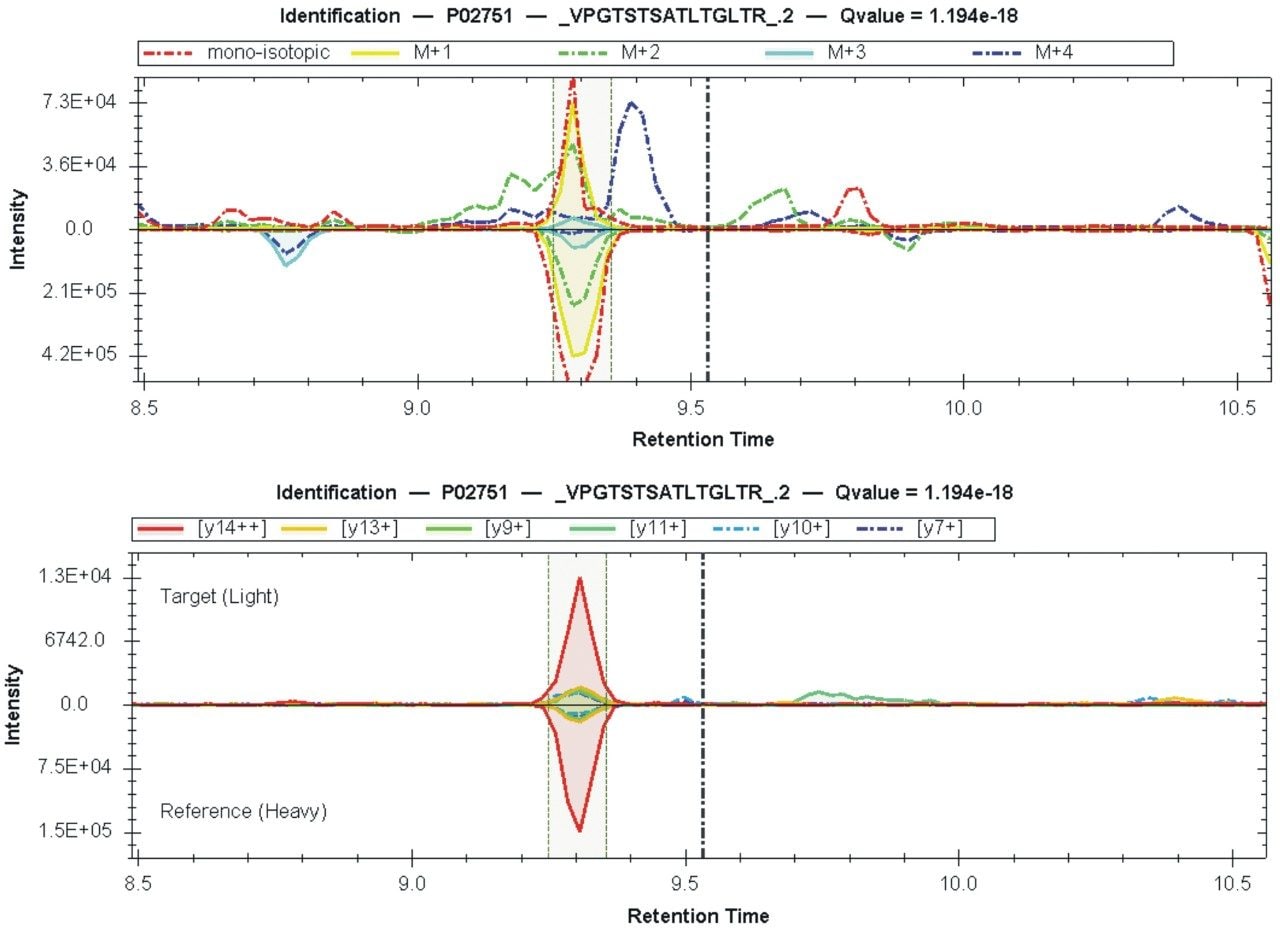
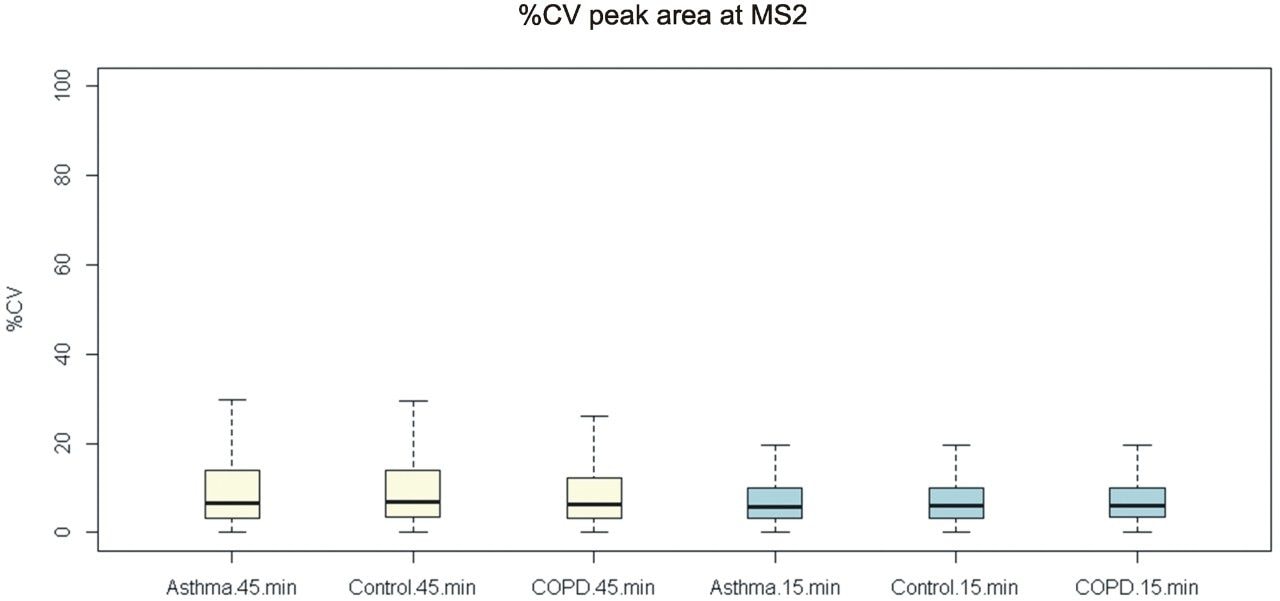
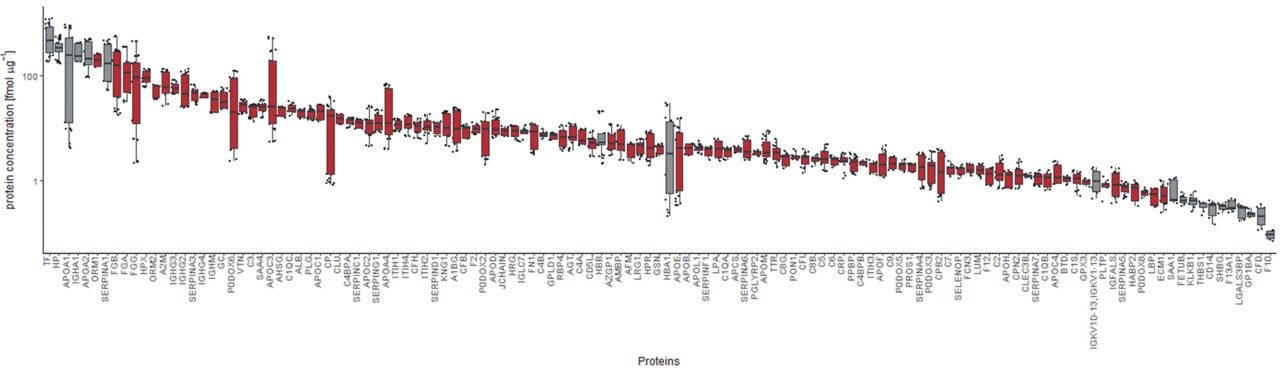
The ability to sufficiently gain a high number of proteins, while achieving high quantitative precision is critical for large cohort, high throughput applications. Implementation of a fast scanning quadrupole, allows this to be achieved. The importance of peak sampling frequency and its effect on quantitative precision is described in more detail elsewhere.2 Figure 4 shows the complexity which is introduced with co-eluting peptides for a 15 min gradient, providing significant opportunities for interference effects. Increased specificity and high rates of data acquisition provided by SONAR, result in non-interfered detection and extraction of MS2 fragment ions allowing proteins to be quantified over a wide dynamic range (>4 orders). The quantitative variance in terms of average %CV within each cohort (over technical replicates) is demonstrated to be 6% for data acquired using a 45 min gradient. Reducing the gradient to 15 min shows the average %CV to be maintained at 6% across the three groups. Data representing the shorter gradient also provides less variance overall, with all measurements falling within 20% CV (Figure 5). Absolute quantitation is provided for proteins corresponding with the PQ500 SIL peptides but also identifications external to the kit (Figure 6).
Overall, results from this study highlight the qualitative and quantitative advantages of SONAR for high throughput proteomics. The implementation of a scanning quadrupole with SONAR acquisitions allow data to be acquired at a faster rate, therefore providing high numbers of protein identifications with higher quantitative precision. A combined workflow with the PQ500 kit demonstrates absolute quantification using the DIA approach, which overcomes the technical challenges provided by traditional, targeted MS schema.
720006388, September 2018