For research use only. Not for use in diagnostic procedures.
This is an Application Brief and does not contain a detailed Experimental section.
This application brief demonstrates analytically sensitive, accurate, and robust quantification of intact IGF-I in biological matrix using a simple sample preparation workflow, an analytical LC and HRMS.
HRMS platforms, with their ability to provide qualitative and quantitative information offer a flexible alternative to TQ systems, especially for direct quantification of intact proteins. With improvements in hardware and software, HRMS platforms can now achieve the performance characteristics required for bioanalytical quantification, as highlighted by the analytically sensitive (10 ng/mL) and robust (CV’s <10%) quantification of intact IGF-I for clinical research.
HRMS – with its versatility, mass accuracy, and high mass resolution – is a well-established tool for qualitative analysis, whereas tandem quadrupole (TQ) mass spectrometers are the gold standard for analytically sensitive and robust quantification. With advances in high resolution mass spectrometry (HRMS) technology, it is becoming an attractive tool for bioanalytical quantification.
Use of biologics (peptides/proteins) as therapeutic agents has increased in the past decade. Historically, these biologics have been quantified using immunoassays. While immunoassays are highly analytically sensitive, they may lack selectivity, standardization, and multiplexing capabilities. With its many benefits (e.g., multiplexing, sensitivity, selectivity, and dynamic range), adoption of LC-MS/MS for quantification of large molecules has increased. Use of tandem (triple) quadrupole (TQ) mass spectrometers for quantification of these molecules, especially intact in biological matrices, can still be challenging. As the size of the protein increases, so does the number of observed precursors. Additionally, fragmentation of the precursors is required to ensure selectivity on TQs. However, this leads to distribution of signal across the multiply charged precursors and across the multiple resulting fragments (Figure 1). As a result, an indirect bottom-up approach, employing enzymatic digestion of the protein and quantification of resulting peptides is commonly used. In contrast, intact quantification removes the need for digestion and maintains molecular integrity. The interest in using HRMS for intact quantification continues to grow and is proving to be a complementary alternative to TQs when selectivity challenged.
Using simple sample preparation, analytical scale LC, and an HRMS system, we demonstrate an analytically sensitive, accurate, and robust quantification of intact IGF-I (7.6 kDa) from plasma with quantitative performance comparable to a TQ system.

The high mass accuracy of HRMS systems can result in reduced mass spectrometry (MS) method development times, even with limited knowledge of the analyte. In this case, using the Xevo G2-XS QTof Mass Spectrometer, we set up a targeted time-of-flight (Tof) MRM (Precursor > Precursor) method with Target Enhancement (TE) for the +7 precursor (1093.716 m/z), using very low collision energy, allowing for passage of precursor ions without fragmentation. This helped increase analytical sensitivity (data not shown) as the signal is not diluted among multiple transitions.
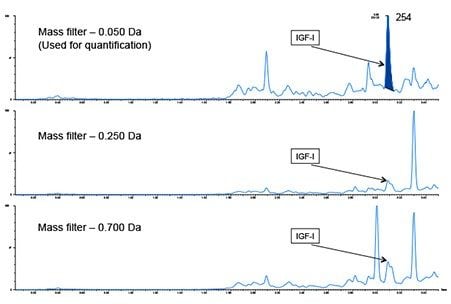
During data processing, different mass tolerances were assessed to determine which would obtain the best quantitative data. As seen in Figure 2, a mass tolerance of 0.050 Da (1093.716 ± 0.050) gave the best results, eliminating interferences and yielding better overall peak areas.
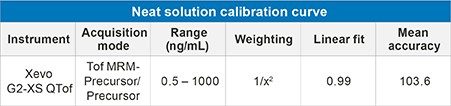
Calibration curve statistics for human IGF-I spiked in neat solution and mouse plasma are highlighted. The neat standard solution calibration curve was linear (1/x2 weighting) over 3.5 orders of magnitude from 0.5–1,000 ng/mL, with a mean accuracy of 103.6% (Table 1).
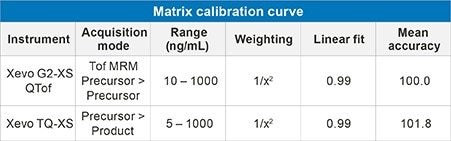
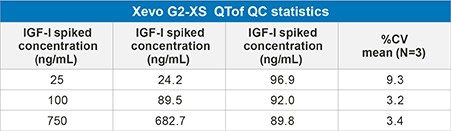
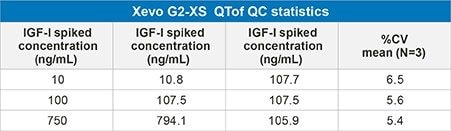
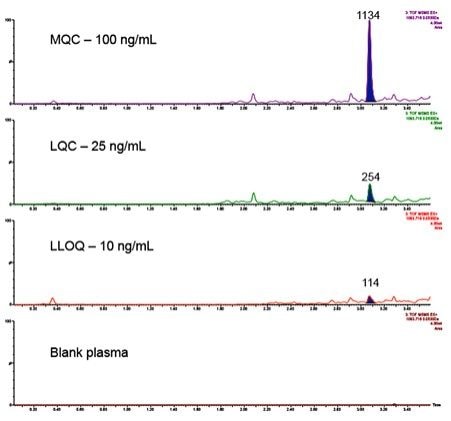
The IGF-I extraction from plasma, employing mixed-mode solid phase extraction (SPE) is detailed in Waters Application Note 720006097EN. In extracted plasma, IGF-I was accurately quantified from 10–1,000 ng/mL. The calibration curve was linear with r2 values > 0.99 (1/x2 weighting) with mean accuracy of all calibration points 100% (Table 2). In addition, QC performance was excellent with mean accuracies between 89-97% and CV’s < 10% (Table 3a, Figure 3). These performance characteristics are comparable to those obtained on the Xevo TQ-XS Mass Spectrometer using MRM mode (Precursor > Fragment) as shown in Table 3b.
HRMS platforms, with their ability to provide qualitative and quantitative information offer a flexible alternative to TQ systems, especially for direct quantification of intact proteins. With improvements in hardware and software, HRMS platforms can now achieve the performance characteristics required for bioanalytical quantification, as highlighted by the analytically sensitive (10 ng/mL) and robust (CV’s <10%) quantification of intact IGF-I for clinical research.
720006167, April 2018