This is an Application Brief and does not contain a detailed Experimental section.
This application brief describes BiopharmaLynx 1.1, a software tool that automates the processing, annotation, and reporting of biotherapeutic protein and peptide map LC-MS data.
Protein-based pharmaceuticals represent an increasing proportion of drug development. These complicated molecules require extensive characterization to achieve regulatory approval, implement manufacturing improvements, and defend intellectual property.
Biopharmaceutical scientists confront many challenges when characterizing and developing therapeutic protein products:
Mass spectrometry plays a critical role in characterizing protein pharmaceuticals. MS may be used to determine the mass and heterogeneity profile of a biotherapeutic protein, confirm the expected primary structure and modification from a peptide map, and probe the tertiary structure through analysis of disulfide bonding patterns.
Analysis, annotation, comparison, and reporting of intact protein and peptide map LC-MS results can be a tedious process that consumes significant time and effort for experienced analysts. Errors resulting from a manual curation process may lead to poor decisions and delayed development timelines for candidate therapeutics. Such errors may even result in selection of candidate clones and proteins with an undesirable safety, efficacy, or process profile. Ideally, data processing analysis tasks should be performed using a standardized methodology that is automated as much as possible, but still permits scientists to use their knowledge to override any incorrect results from the automated analysis. Perhaps the most common workflow is to directly compare data from a “gold standard” reference material to a batch of test samples to identify new/missing components or components that have changed in intensity. Software that facilitates these comparisons has been long desired to improve productivity of analytical groups in biopharmaceutical organizations.
Waters BiopharmaLynx 1.1 automates the processing, annotation, and reporting of biotherapeutic protein and peptide map LC-MS data sets for maximum laboratory productivity. BiopharmaLynx matches chromatographic peaks in a map or spectrum with structural features of proteins, allowing peptides, proteins, and their variants to be detected. Advanced tools for visualizing results and comparing results between samples have been included, so that sample differences may be readily determined and communicated.
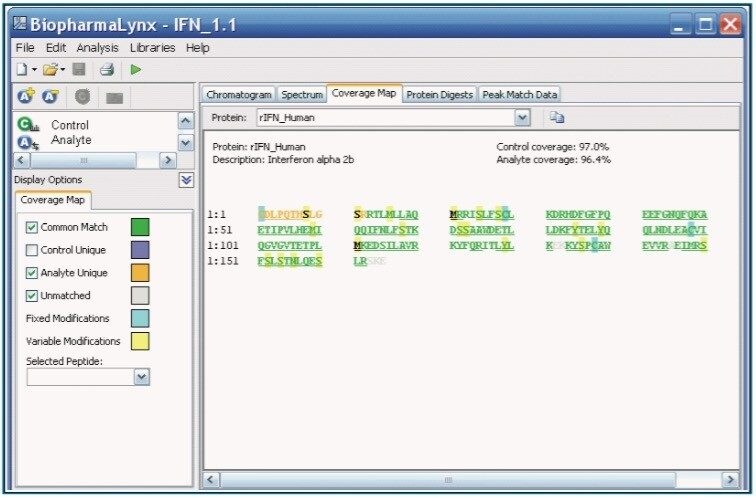
Peptide mapping provides information on the primary structure of a protein, localizes modifications to specific peptides, and confirms tertiary structural elements, such as disulfide bond patterns. Furthermore, for modifications with small or no mass differences (e.g. deamidation or isomeric glycan structures), the combination of high resolution UltraPerformance LC (UPLC) separations with accurate mass determinations are essential investigatory tools.
BiopharmaLynx 1.1 has been optimized for LC- time-of-flight (TOF)-MS peptide map analysis, and is capable of recognizing and annotating peptide ions that arise from fully-digested, modified, and semi-digested proteins (e.g. missed cleavages and semitryptic peptides). In addition, the capability to recognize sequence confirming in-source fragments has been enabled in BiopharmaLynx 1.1.
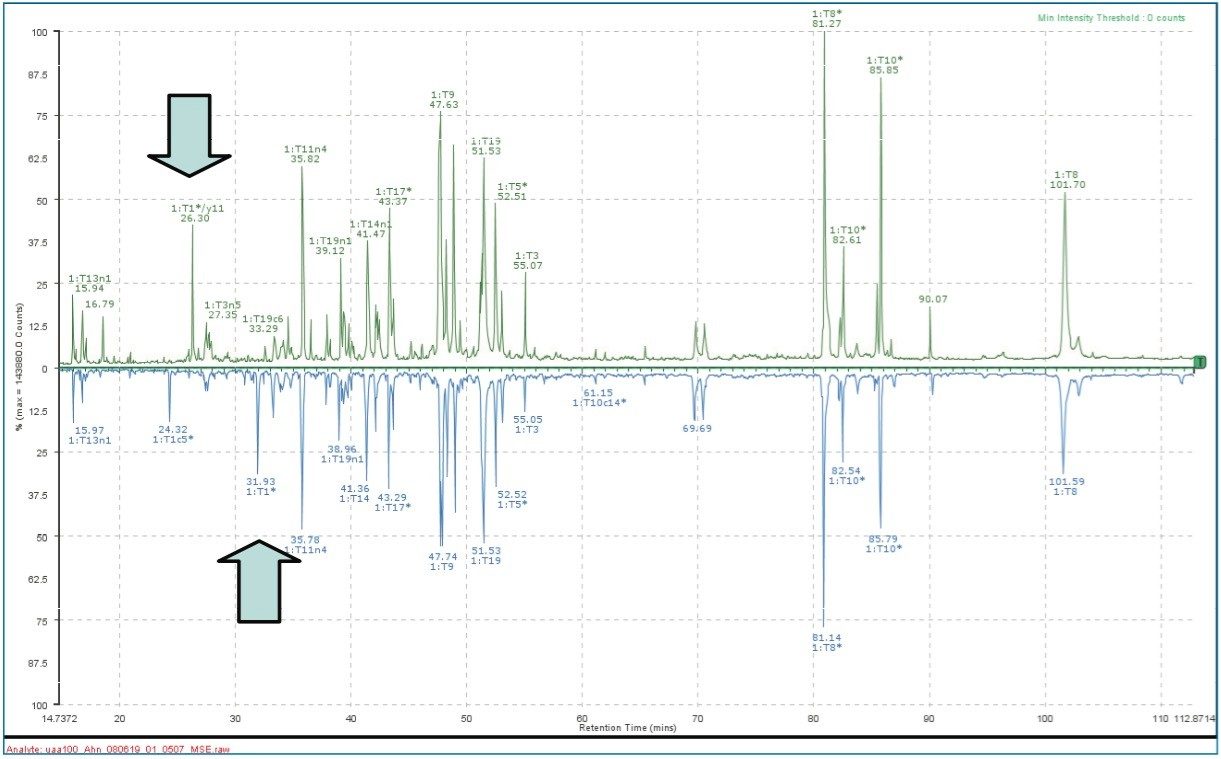
For peptide mapping analysis, BiopharmaLynx 1.1 features:
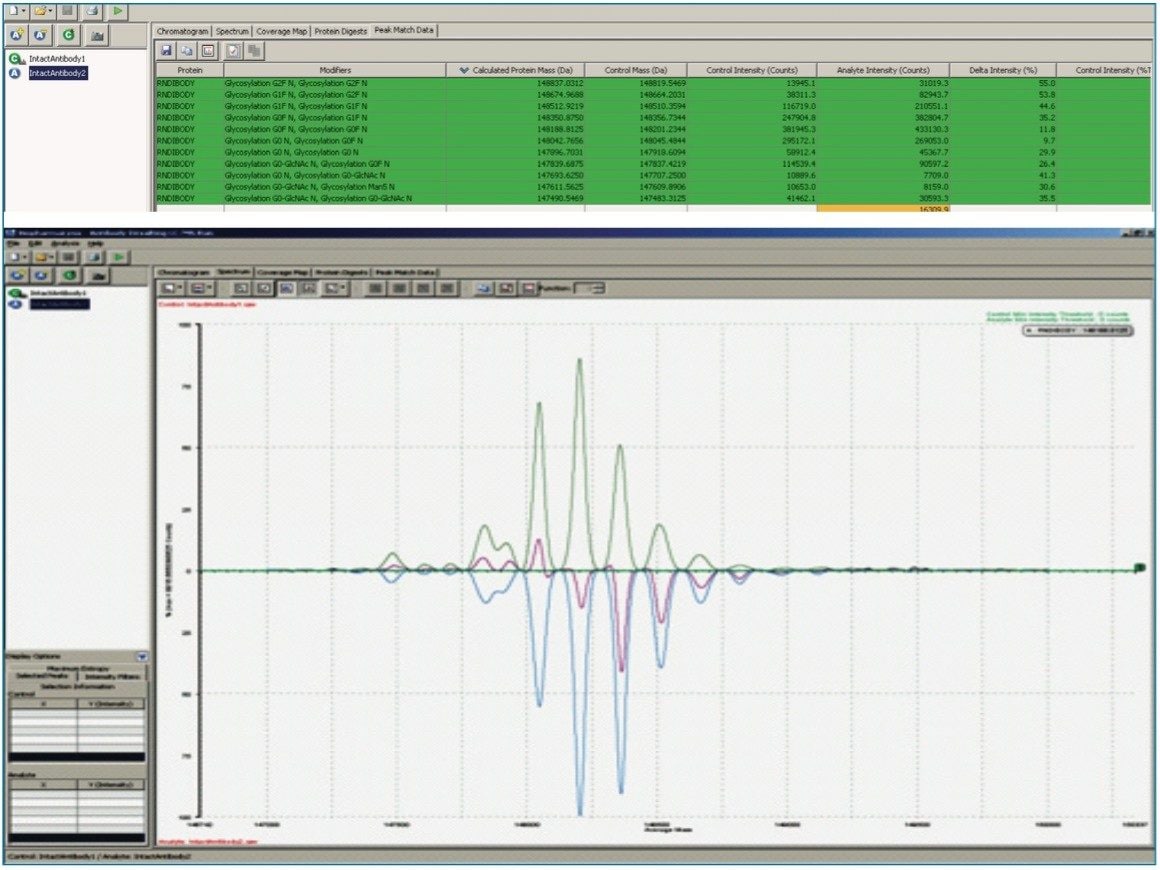
Intact mass analysis using TOF MS provides an accurate molecular weight and a fingerprint of the protein’s heterogeneity. Patterns of glycosylation, protein processing, and modification represent a holistic view of a biotherapeutic, which may be used to quickly assess process and batch variation.
Proteins produce a population of multiply-charged ions during electrospray ionization that must be deconvoluted for accurate mass determinations. BiopharmaLynx performs spectral deconvolution using the latest generation of the MaxEnt1 maximum entropy deconvolution algorithm, which has been used to facilitate protein mass determinations in academia and the pharmaceutical industry for over 15 years. Results are reproducible, show superior resolution, and maintain relative component quantitation compared with the raw mass spectral data.
720002794, October 2008