In this application note, a rapid method for analysis of glycans is presented. This method describes deglycosylation of glycoproteins aided by an enzyme-friendly surfactant (RapiGest SF), sample cleanup using a HILIC chromatography performed in a 96-well microtiter plate (MassPREP HILIC µElution Plate), and MALDI MS analysis of the resulting glycans.
Glycosylation is one of the most important types of posttranslational modification (PTM) in proteins. Due to the high degree of heterogeneity, the characterization of glycans is a challenging task. Mass spectrometry (MS) is a primary tool for biopolymer analysis; however, the characterization of (native) glycans is complicated by the time-consuming sample preparation and their poor MS ionization efficiency. A typical sample preparation method for MS involves a chemical or enzymatic cleavage of glycans, followed by salts, surfactants, and protein residues removal. Purified native glycans can be directly analyzed by MALDI-Tof MS.
The efficient sample deglycosylation is a key requirement for a successful and sensitive glycan analysis. Nevertheless, the quantitative glycan release (e.g., using enzymes) is rarely achieved, since the glycosylated sites of the proteins are often obstructed by the protein secondaryband tertiary structure.
The goal of this work was to develop a rapid and efficient deglycosylation of N-linked glyco proteins with a glycosidase (PNGase F) aided with the enzyme-friendly surfactant, RapiGest SF. This was followed with a novel micro-scale hydrophilic-interaction chromatography (HILIC) solid-phase extraction (SPE) plate (Waters MassPREP HILIC µElution Plate) for a rapid sample cleanup prior to MALDI MS analysis using highly purified MALDI matrix (Waters MassPREP MALDI Matrix, DHB).
The glyco proteins were solubilized in 0.1% (w/v) RapiGest SF solution prepared in 50 mM NH4HCO3 buffer, pH 7.9. Protein samples (e.g., ovalbumin) were reduced with 10 mM DTT for 45 min at 56 °C and alkylated with 20 mM iodoacetamide in the dark for 1 h at room temperature. The enzyme PNGase F (2.5–5 units) was added, and the protein solutions were incubated for 2 h at 37 °C.
The RP HPLC instrument (CapLC XE, Waters) was equipped with a microbore RP-HPLC column (Waters Atlantis dC18 Column, 3.5 mm, 1.0 x 100 mm). The LC separation was hyphenated with a Waters Micromass Q-Tof micro. Mobile phase A was made of 0.1% formic acid in Milli - Q water (Millipore Corp., Billerica. Massachusetts). Mobile phase B was made of 0.1% formic acid in 100% acetonitrile. A linear gradient was run from 0 to 60% B in 30 min (2% B per min). Separation was carried out with 35 mL/min flow rate; the column temperature was set at 40 °C.
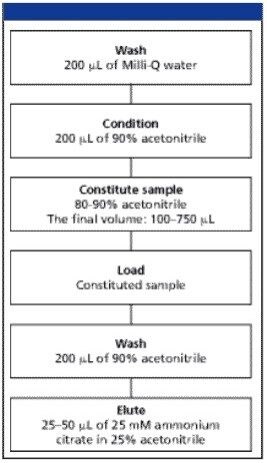
The N-linked glycans released from glyco proteins were extracted using the 96-well, MassPREP HILIC µElution Plate attached to a vacuum manifold. Use of this SPE device involves an initial wash and equilibration of the sample well(s), sample loading, sample well washing to remove undesired products, and final elution of the isolated glycans. Figure 1 shows the optimized MassPREP HILIC µElution Plate SPE protocol for both neutral and the sialylated glycans. The entire process requires less than 20 min. The HILIC plate performance was evaluated with maltoheptaose standard. Load, wash, and elution SPE fractions we re quantitatively analyzed by an HPLC system with evaporative light scattering detection (ELSD). The mass balance revealed no break through in the load fraction. Most of the material eluted in the first 25 mL elution. Total mass balance was 90%. Recovery was estimated to be approximately 70%.
Ultra pure MassPREP MALDI matrix, DHB (2,5-Dihydroxybenzoic acid) was used for MALDI-Tof analysis. The matrix was reconstituted in 500 mL of pure ethanol to a final concentration of 20 mg/mL. Purified glycan solutions were mixed with DHB matrix in one to one ratio; one mL was placed onto a stainless steel MALDI target. Waters Q-Tof Ultima MALDI was used to determine the molecular weight of the released glycans and perform MS-MS experiments to characterize the structure of the glycans. The typical collision energy used here was 70 to 120 V.
In earlier reports, we described the use of a mild and enzyme-friendly surfactant, RapiGest SF, for denaturation of proteins prior to proteolytic enzymatic digestions.1 It was found that this surfactant improves the speed and completeness of enzymatic proteolysis, most noticeably for globular and membrane proteins.2 Therefore, we investigated the use of RapiGest SF in conjunction with PNGase F for the enzymatic release of N-linked glycans.
Figure 2 shows the extent of the deglycosylation reaction of chicken ovalbumin solubilized in 0.1% RapiGest SF (Figure 2c) digested with PNGase F for 2 h in 50 mM ammonium bicarbonate solution. The pro g ress of deglycosylation is apparent in comparison to a control ovalbumin sample (Figure 2a) with no enzyme added. The deglycosylation was also carried out with the addition of 0.1% of non-ionic surfactant, n-octyl-β-glycopyranoside (OG) (Figure 2b).
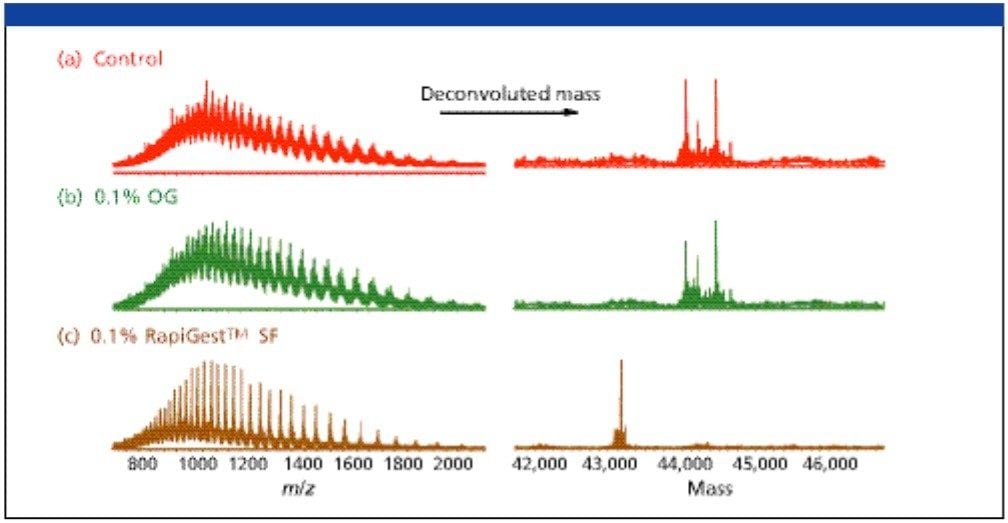
The LC-MS analysis of samples produced the ESI spectra featuring the multiply charged protein states, which were deconvoluted using Waters Micromass MassLynx MaxEnt 1 software (deconvoluted MS spectra are shown in the right panel in Figure 2). As expected, no signal corresponding to the MW of deglycosylated p rotein was found in the control sample (Figure 2a). Interestingly, no distinguishable deglycosylation was also observed in the OG-mediated deglycosylation (Figure 2b). Multiple peaks between 44–45 kDa re p resent the various N-linked glycoforms of ovalbumin. The reaction in the presence of RapiGest SF shows nearly complete deglycosylation; the protein mass shifted and a prominent peak was detected at approximately 43 Kda, which is consistent with the MW of the unmodified protein.
The glycans were extracted using the MassPREP HILIC µElution plate. In a HILIC mode, the hydrophilic glycans are retained due to a partitioning separation mechanism between the organic mobile phase and a layer of water adsorbed on the surface of sorbent. Since the high concentration of organic solvent is necessary to ensure good retention of glycans, the samples were first diluted with ACN to a final concentration of 80–90%. Some precipitation of glycans may occur if they are present at high concentrations. It is not recommended to centrifuge samples prior to loading to the HILIC µElution plate. After plate conditioning (sample cleanup section in experimental), glycan samples were loaded by gravity (Figure 1).
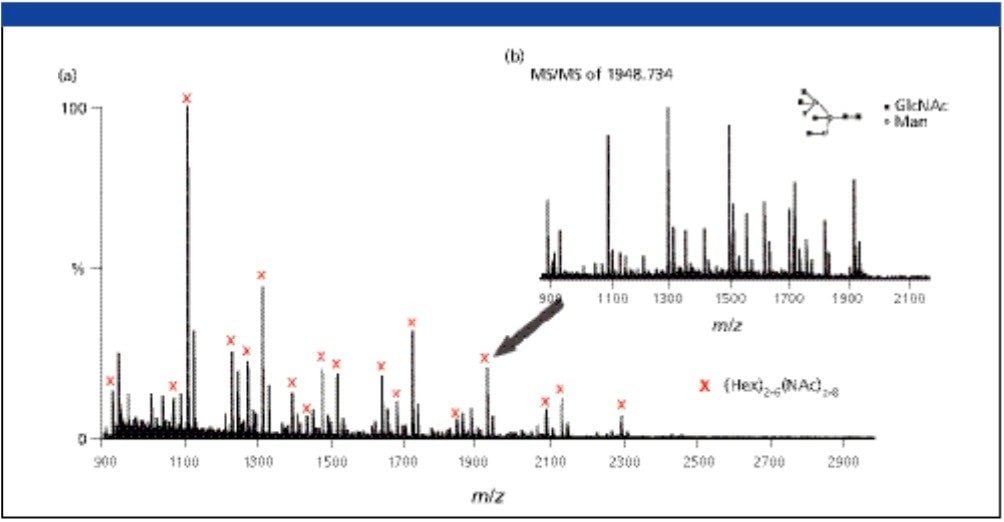
The MALDI Q-Tof MS spectra of underivatized N-linked glycans released from 10 pmol Ovalbumin were obtained (Figure 3). MS-MS fragmentations of selected ions were performed to validate the glycan structures. For example, collision induced dissociation of the complex glycan ion of mass to charge ratio of 1948.734 (M + Na) was shown (Figure 3). This ion is observed in the MS mode with low ion intensity, however, enough fragmentation ions were produced in the MS-MS mode to determine its structure (GlycoSuite database, Proteome Systems, Ltd.).
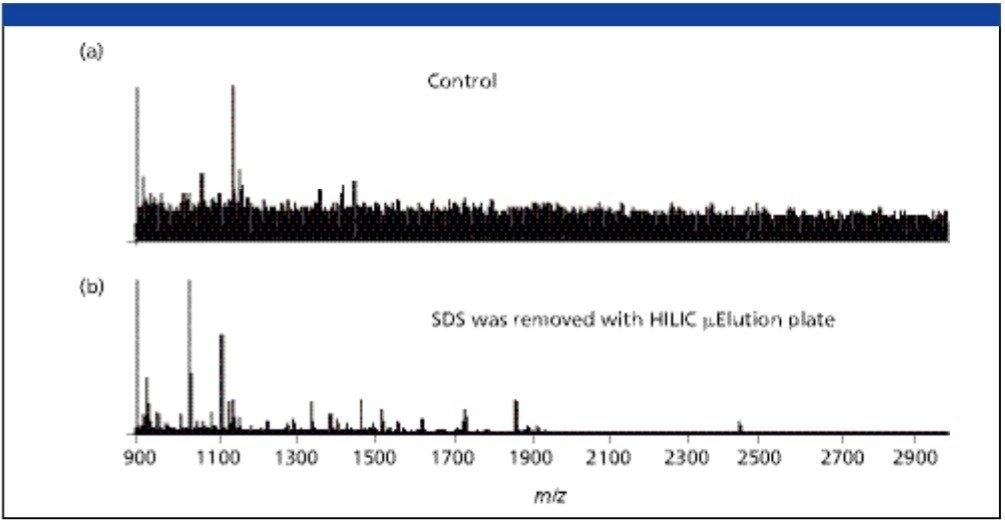
The MassPREP HILIC μElution Plate facilitates the removal of impurities including the surfactants, such as RapiGest SF from the sample. It can be used for surfactant removal in general, for example SDS from peptides/glycopeptides. Figure 4 shows the MALDI MS analysis of the bovine serum albumin (BSA) tryptic digest. No signal was observed for the sample contaminated with 0.1% SDS, while BSA tryptic peptide signals we re observed in high abundance without any ion suppression caused by the presence SDS.
We have developed a method suitable for fast and robust analysis of glycans released from glyco proteins. The method utilizes an enzyme friendly surfactant (RapiGest SF) that was shown to greatly accelerate a deglycosylation reaction via glyco protein denaturation, which makes the glycans more accessible to enzymatic cleavage. A complete deglycosylation of proteins was achieved after 2 hr incubation with PNGase F. The MassPREP HILIC µElution plate was utilized to extract and desalt the glycans prior to their MS analysis using MassPREP MALDI Matrix, DHB. The SPE method is fast and requires minimum sample manipulation.
WA41849, June 2007