Application Brief
This is an Application Brief and does not contain a detailed Experimental section.
Szabolcs Fekete, Mateusz Imiolek
1 Waters Corporation, Geneva, Switzerland
Published on May 26, 2025
This is an Application Brief and does not contain a detailed Experimental section.
Programmed injection techniques, including sequential and bracketed injection modes, are increasingly important tools for improving chromatographic workflows. These advanced injection strategies are particularly valuable for minimizing solvent mismatch, reducing band broadening, enhancing peak symmetry, and automating sample preparation, especially when analyzing large biomolecules such as monoclonal antibodies (mAbs), nucleic acids, and lipid nanoparticles (LNPs). Empower Chromatographic Data System (CDS) offers flexible implementations — such as Auto Additions and In-Needle Auto Additions — to realize these advanced injection sequences in an automated and reproducible manner. This application note offers practical guide to performing programmed injections in Empower CDS, highlighting how these methods can optimize sample introduction and chromatographic performance in challenging applications.
Bracketed or sequential (also referred to as stacked or programmed) injection modes are particularly valuable for the analysis of complex biotherapeutics.1 Modern autosamplers are increasingly capable of performing sophisticated liquid handling tasks beyond simple sample injection, including mixing, dilution, derivatization and internal standard addition.2,3 Among these capabilities, sequential injection methods or performance-optimized injection sequence (POISe) - have been shown to provide significant analytical benefits by minimizing solvent mismatch effects and reducing extra-column band broadening.4,5,6 POISe techniques, in which weak solvent plugs are co-injected with the sample, improve solute focusing (at the head of the column), resulting in sharper peaks and mitigating band broadening. Despite these advantages, these approaches remain underutilized in routine chromatographic workflows.
Various programmed injection strategies have demonstrated their usefulness in a variety of biopharmaceutical applications. In anion exchange chromatography (AEX) of large nucleic acids, such as mRNA, a sequential injection using a high salt concentration bracketing plug (i.e. injecting nucleic acids between plugs of 2 M NaBr at pH ~10) significantly reduced carryover and improved analyte recovery.7 Similarly, hydrophilic interaction chromatography (HILIC) analyses of monoclonal antibodies (mAbs) can benefit from sequential injection methods. A mismatch between the aqueous sample diluent and the highly organic mobile phase can lead to solute breakthrough and distorted peak shapes.8,9 "Bracketing" the injection of aqueous mAb samples with acetonitrile (MeCN) plugs has been shown to mitigate this mismatch, allowing injection of larger sample volumes (up to 5% of the column volume) without compromising peak symmetry or inducing breakthrough.10 In addition, programmed injection strategies facilitate sample preparation for size exclusion chromatography (SEC) of lipid nanoparticles (LNPs) encapsulating nucleic acids. LNP formulations are sensitive to mechanical and chemical stress and require gentle and reproducible handling. In this context, automated sequential injection programs have been used to dilute LNP samples and introduce detergents to release encapsulated RNA payloads.11 All those automated injection methods improve reproducibility, reduce human error and limit analyst exposure to hazardous reagents.
In summary, programmed (bracketed, sequenced or stacked) injection modes represent a highly effective approach to improving the chromatographic performance or sample preparation of large molecule biotherapeutics.
This application note provides a brief technical description of how to implement programmed injections in the Empower Chromatographic Data System (CDS).
1. Prep Type / In-Needle Auto Additions / Sample Prep (Empower CDS 3.6.0 and above)
In later versions of Empower CDS (3.6.0 and above) you can select the Prep Type column in the Sample Set table and use the In-Needle Auto Additions function to access advanced injection and sample preparation options.12,13 If the Prep Type and In-Needle Auto Additions columns do not appear in the Sample Set, the Prep Type and Sample Prep columns must be enabled (“unhidden”) in the Table Properties to make it visible and accessible. You can do this by going to the Table Properties (of the Sample Set) and checking the status of Columns and Hidden Columns (Figure 1). Once the Prep Type and Sample Prep are accessible, it is easy to program injection sequences.
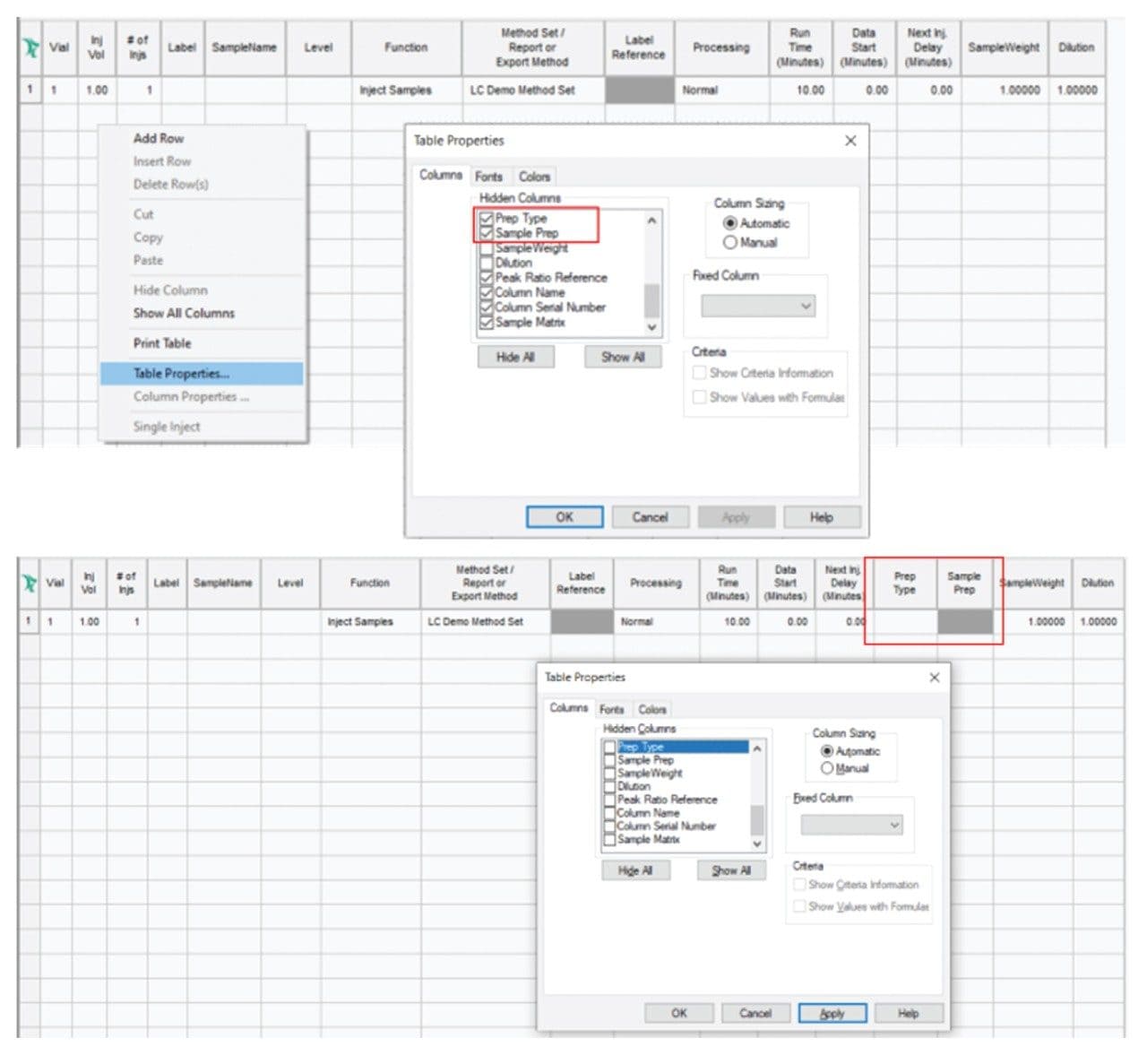
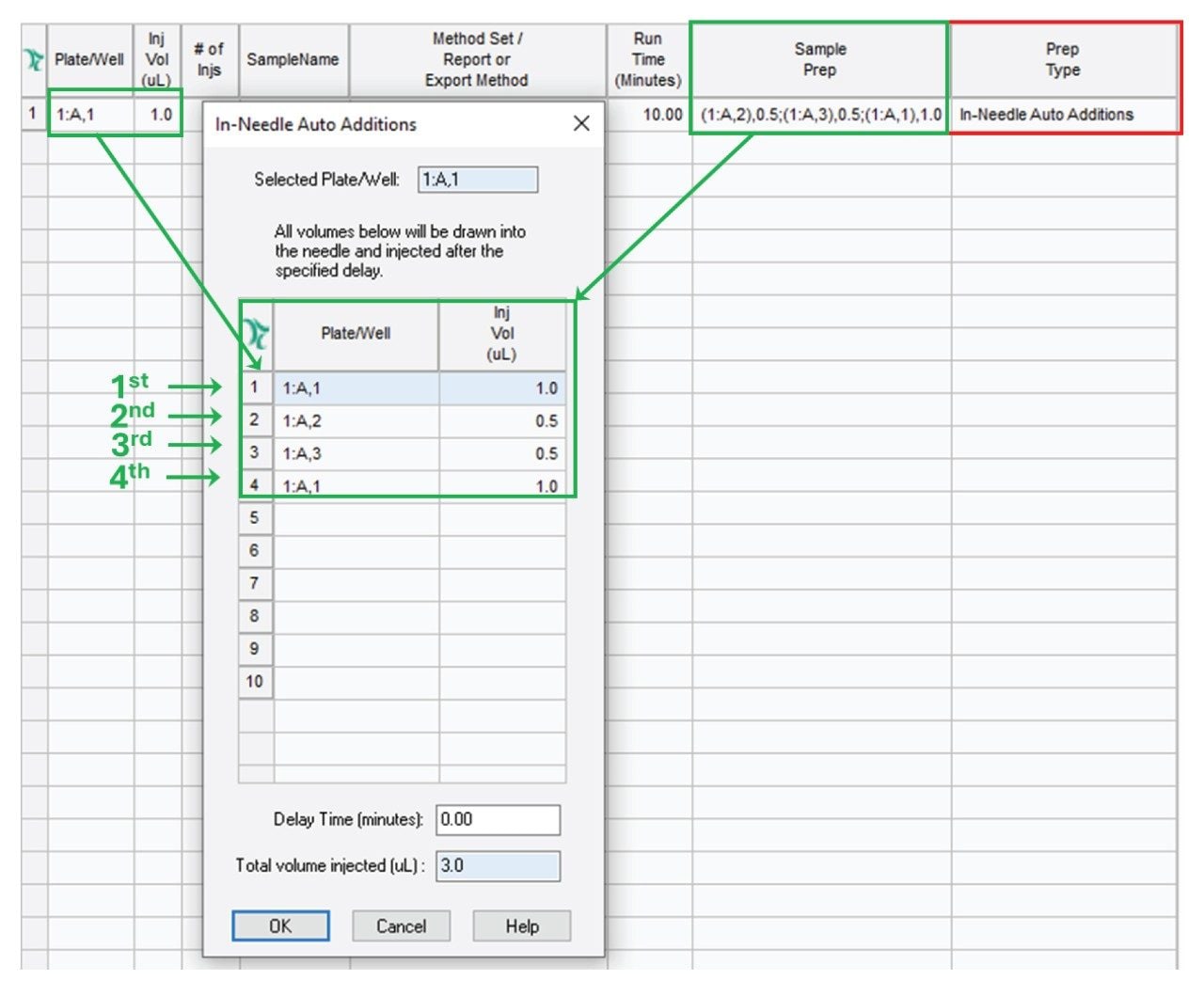
Consider the following example: We have a solvent in vial 1:A,1 and two separate samples in vials 1:A,2 and 1:A,3. We are going to mix the two samples and introduce the mixture, bracketed in a solvent plug, into the flow path. Figure 2 shows the appropriate setting for In-Needle Auto Additions. First 1.0 µL is drawn from 1:A,1 (solvent), then 0.5 µL from vials 1:A,2 and 1:A,3 and finally 1.0 µL of solvent is drawn from 1:A,1. With such an injection sequence, the needle will inject the entire 3 µL (1+0.5+0.5+1 µL) of liquid.
2. Auto Additions (Empower CDS up to the version 3.6.0)
In earlier versions of Empower CDS (up to the version 3.6.0), the injection and sample preparation options are accessible via the Auto Additions option in the Sample Set.12,13 If the Auto Additions column does not appear in the Sample Set, the Auto Additions column must be enabled (“unhidden”) in the Table Properties to make it visible and accessible. You can do this by going to the Table Properties (of the Sample Set) and checking the status of Columns and Hidden Columns. Once the Auto Additions is accessible, it is easy to program injection sequences.
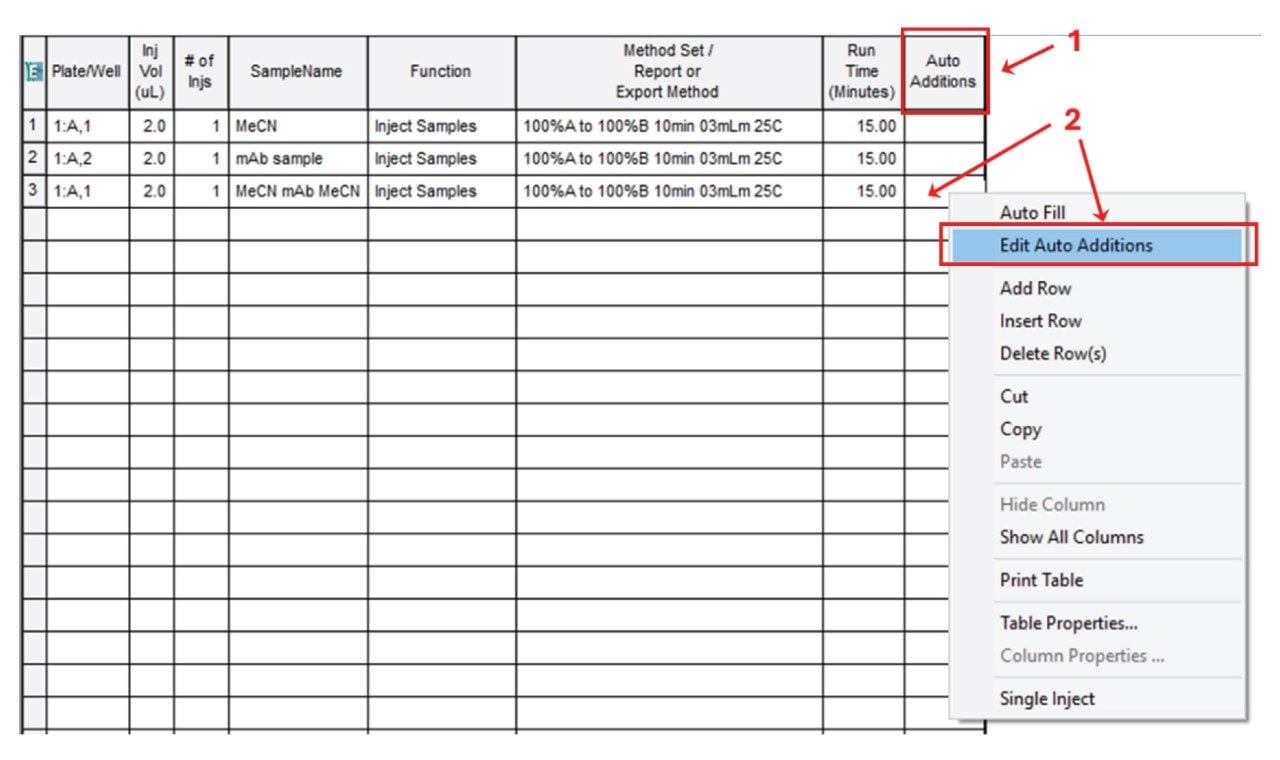
Consider the following example: We have acetonitrile (MeCN) in the vial 1:A,1 and a monoclonal antibody (mAb) sample in the vial 1:A,2 and we are going to perform a bracketed injection for a HILIC separation. Figure 3 shows the corresponding Sample Set. We plan to first perform a MeCN blank injection (line 1 in the Sample Set), then a mAb sample injection (line 2 in the Sample Set) and finally we “bracket” the mAb sample between MeCN solvent plugs (line 3 in the Sample Set).
To perform the bracketed injection we need to indicate the position of the first vial (MeCN) - of the sequence - in the Plate/Well column of the Sample Set. Figure 3 shows that a 2.0 µL volume is first taken from the vial placed at the 1:A,1 position. Then click on the Auto Additions cell and select Edit Auto Additions from the drop-down menu (shown in Figure 3).
This opens the Auto Additions Editor where you can program the injection series by specifying the position of the vials to be drawn from and the corresponding volumes (Figure 4). In our example, 1:A,2 (mAb sample) and 1:A,1 (MeCN) are both specified with volumes of 2.0 µL. The final sequence is as follows: First the needle takes 2.0 µL from 1:A,1, then 2.0 µL from 1:A,2 and finally 2.0 µL again from 1:A,1 (Figure 4). The user can complete the command by clicking the OK button in the Auto Additions Editor. With such an injection sequence, the needle will inject the entire 6 µL (2+2+2 µL) liquid volume into the flow path.
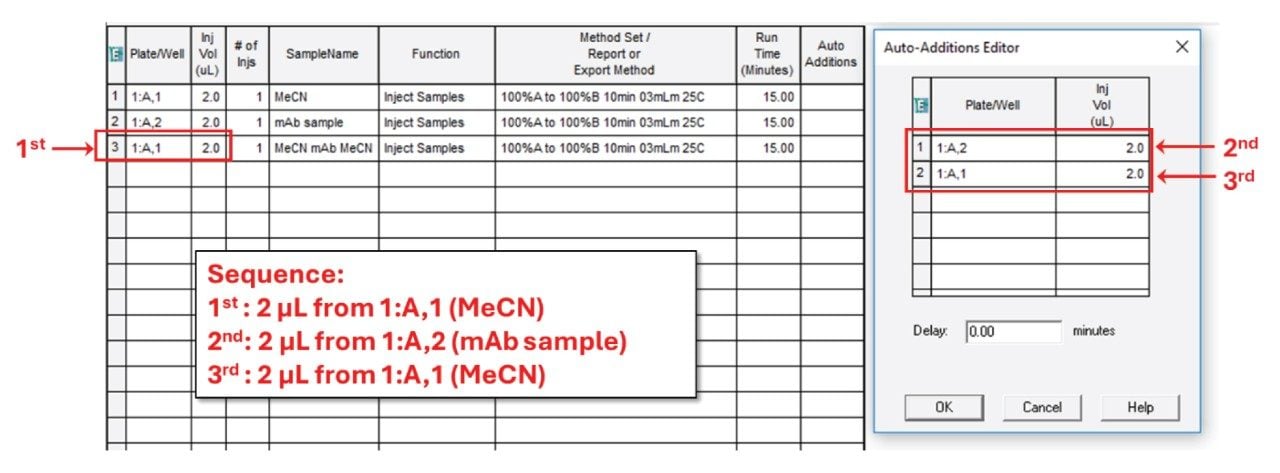
Once the commands in the Auto Additions Editor have been completed, the positions of the vials included in a given injection sequence and the corresponding volumes appear in the Auto Additions cell of the Sample Set (Figure 5).

Note the difference in drawing order between Auto Additions and Sample Prep's In-Needle Auto Additions. The former does not list the entire drawing/injecting sequence in its editor. The first draw is indicated in the Plate/Well cell of the Sample Set and the Auto Additions Editor lists the rest of the sequence events (from the second draw). In contrast, the Sample Prep In-Needle Auto Additions editor lists the entire injection sequence.
3. Example
The example in Figure 6 compares a simple injection of an oligonucleotide (ON) ladder in HILIC mode (panel A) and a programmed injection sequence where the ON sample is bracketed with MeCN solvent plugs (panel B). The simple injection results in a severe breakthrough effect due to solvent mismatch (signal increase at column dead time) and splitting for peaks 1, 2, and 3. In contrast, when the ON sample is injected between weak solvent plugs (MeCN), no splitting or breakthrough is observed.
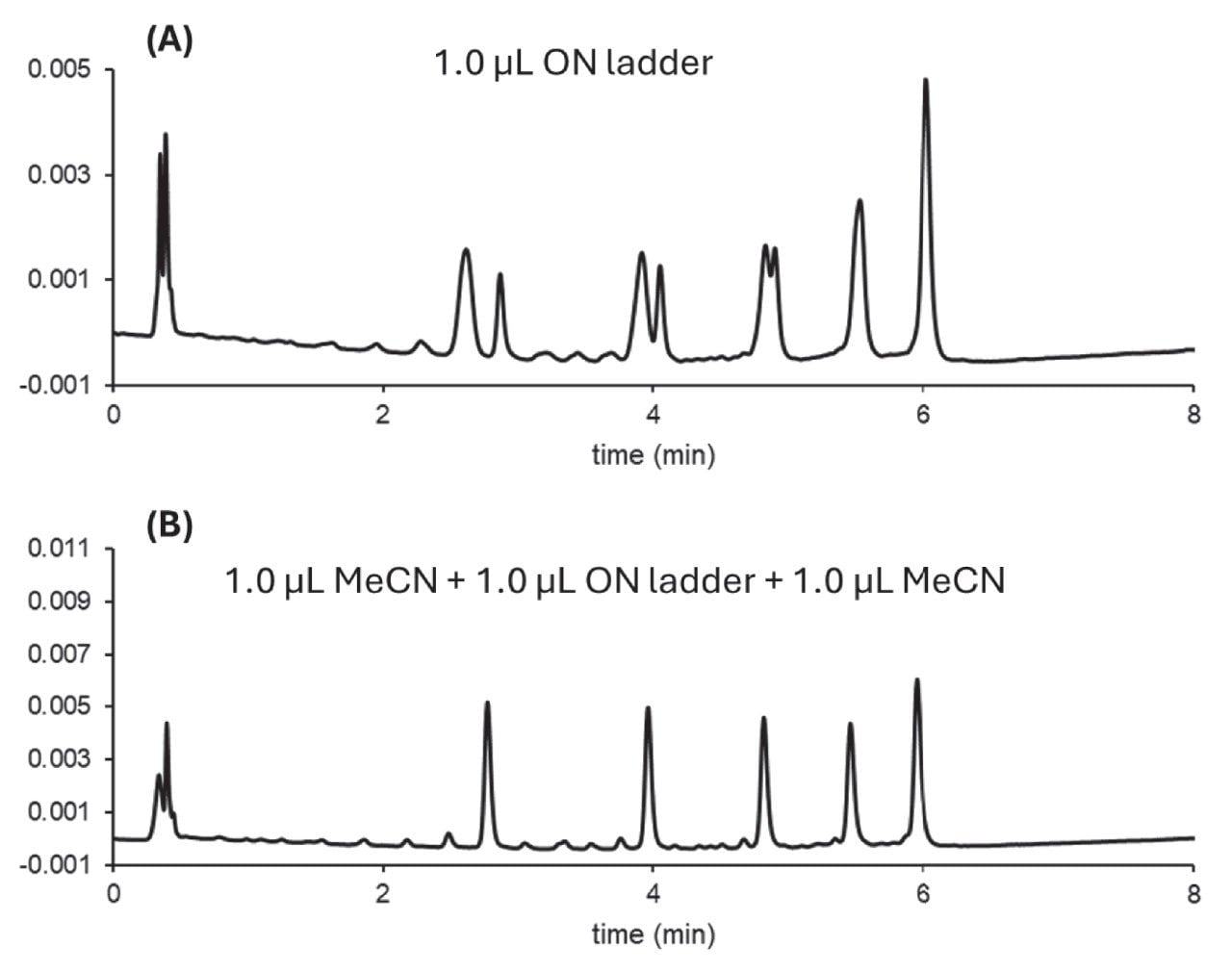
Programmed injection techniques are highly effective for overcoming sample introduction challenges associated with the chromatographic analysis of biotherapeutics. By leveraging Empower CDS capabilities such as Auto Additions and In-Needle Auto Additions, users can automate advanced injection sequences. This not only improves analytical performance but also enhances workflow efficiency and reproducibility. Empower CDS provides an accessible and robust platform to implement these techniques, supporting the evolving needs of biopharmaceutical analysis.
720008786, May 2025