High-Resolution Size Exclusion Chromatography of Megadalton-Sized DNA Vectors and Plasmids Using Waters GTxResolve 2000 Å SEC Columns
Abraham S. Finny, Christian Reidy, Balasubrahmanyam Addepalli, Matthew A. Lauber
Waters Corporation, United States
Published on June 06, 2025
Abstract
Accurate characterization of cell and gene therapy (CGT) drug products is critical for their successful development and deployment. Given their megadalton-sized cargos, these products require carefully optimized chromatographic tools to keep their integrity and native shape for size-based separations. Size exclusion chromatography (SEC) is a preferred technique for this purpose, but its success depends on the use of optimal pore size particles packed in inert hardware to resolve structurally complex samples. Robust SEC methods must also demonstrate high reproducibility, minimal analyte adsorption, and low secondary interactions between analytes and the column hardware or packing materials.
In this application note, the benefits of Waters GTxResolve™ 2000 Å SEC Columns are highlighted, which feature tailored pore and particle sizes specifically designed for the analysis of megadalton-sized molecules. These columns deliver high efficiency, excellent inertness, and superior resolution, recovery, and reproducibility at both the batch-to-batch and column-to-column levels, using standard mobile phases. This represents a significant advancement in performance and reliability compared to conventional 5 µm, 2000 Å pore size columns. Using a DNA vector mixture consisting of plasmid and bacteriophage DNA, as well as thyroglobulin as analytical surrogates, the ability to resolve low-level aggregate species and differentiate molecules based on their structural properties are demonstrated. Importantly, the reproducible chromatographic behavior observed across multiple columns and packing material batches supports faster and more confident assessment of process- and product-related impurities in CGT workflows as well as the plasmids used in all types of recombinant expression. The adoption of these columns for analyzing megadalton-sized biomolecules can help accelerate the development of safer, more efficacious, and globally accessible medicines.
Benefits
- High-resolution megadalton-sized DNA and protein species are separated with clear differentiation of aggregates, monomers, and structural variants.
- Exceptional reproducibility is achieved across columns and packing material batches, with low variability in key chromatographic performance metrics.
- Optimized surface chemistry minimized secondary interactions, enhancing peak symmetry, and analytical robustness.
- Reliable platform for diverse biomolecules demonstrated consistent performance for both nucleic acids and high-molecular-weight proteins.
Introduction
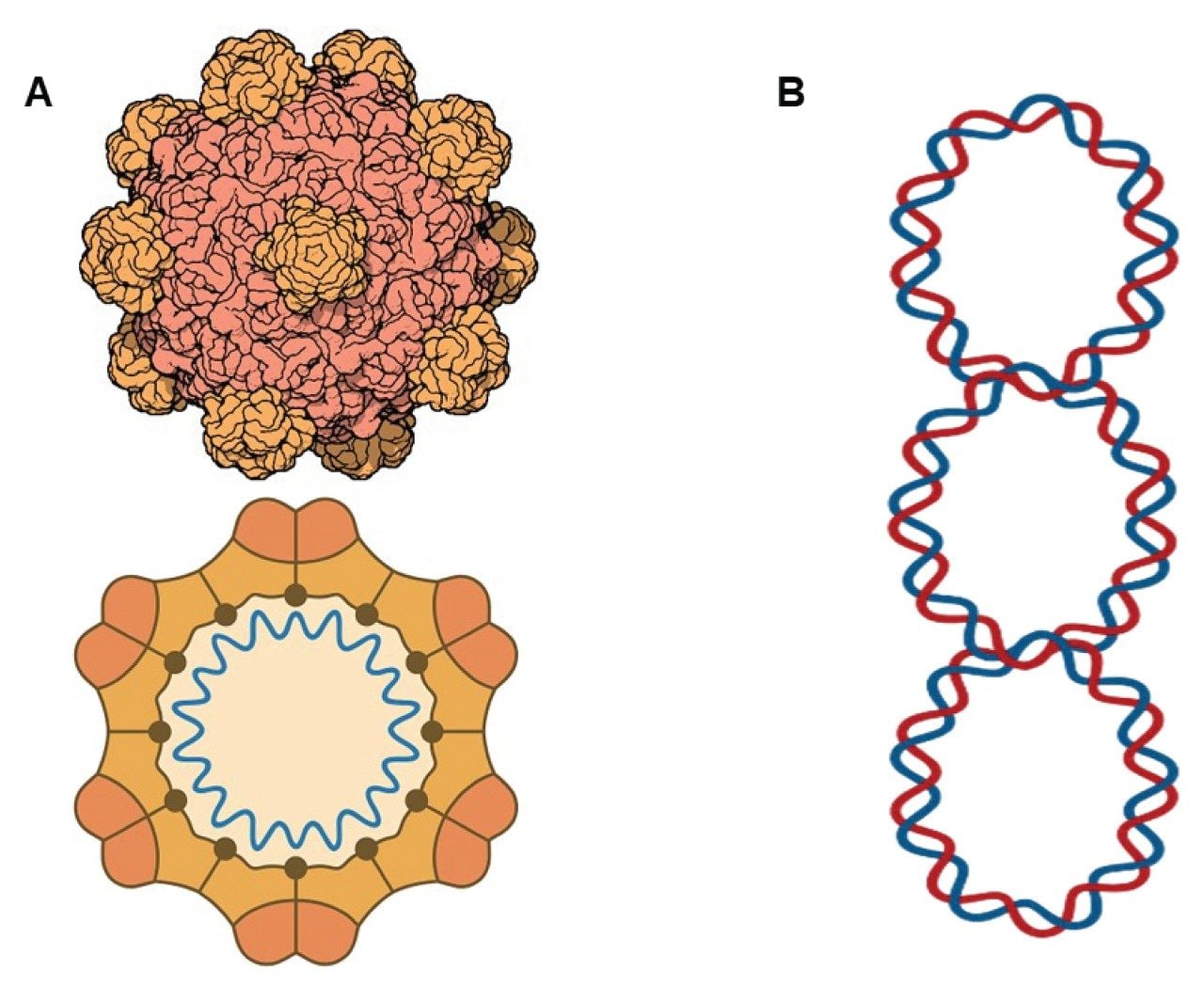
The rise of genetic medicine has led to a new generation of complex modalities ranging from plasmid DNA, mRNA drugs, lipid nanoparticles, and viral vectors with genome editing or drug-related payloads1, 2, 3. These molecules vary widely in their size, structure, and physicochemical properties, and their successful development hinges on precise, reproducible analytical methods that can assess critical quality attributes (CQAs) such as concentration, purity, structural integrity, size distribution, and presence of aggregates. Among these modalities, plasmid DNA plays a central and foundational role. Plasmids are circular double-stranded DNA constructs widely used as expression vectors in recombinant protein production, gene therapy, vaccine development, and genome editing workflows. In the biopharmaceutical industry, plasmids serve as essential starting materials for the manufacture of viral vectors (e.g., AAV, lentivirus), mRNA-based therapeutics, and CRISPR delivery systems. Their quality directly impacts the yield, consistency, and safety of downstream products, making their analytical characterization a key priority in both upstream process development and final product release.
SEC is a robust chromatographic technique that separates analytes based on the relative size or hydrodynamic volume in accordance with the average pore size of the packing material. As a widely accepted and well-established separation technique, SEC has played a significant role in characterizing CQAs of biologic drug substances ranging from peptides to monoclonal antibodies (mAbs) and antibody drug conjugates. Analysis of CGT products, however, requires extra considerations on pore size, pore volume, inert particle surfaces, and column hardware to minimize secondary interactions. Further, there is a need for high-throughput analytical testing, requiring the development of more efficient, smaller particle size packing materials.
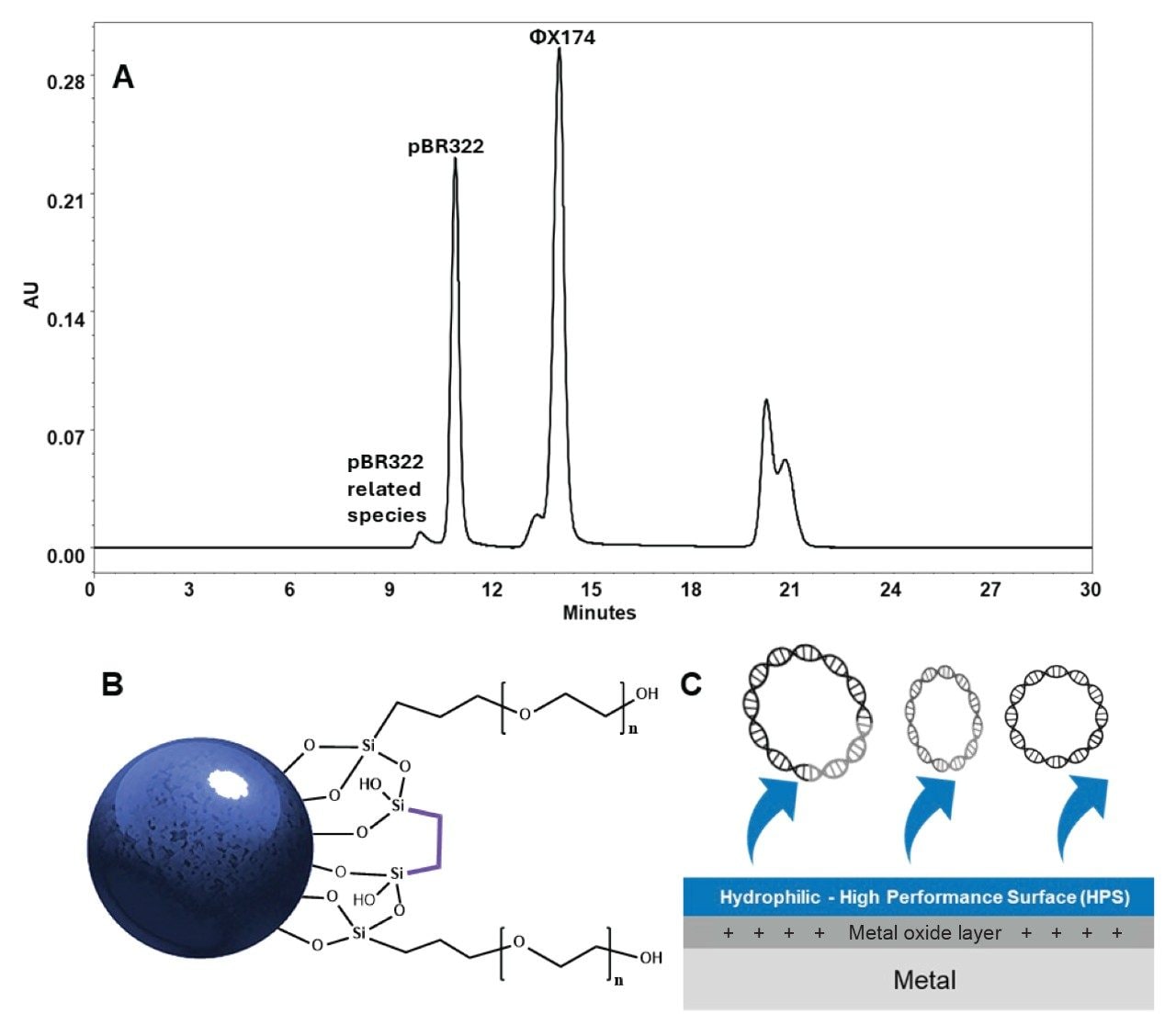
Mega-size biomolecules are highly susceptible to nonideal secondary interactions as a result of their concentrated electrostatic regions and hydrophobic patches. The nonideal interactions mediated by these regions can cause shifts in retention times, poor peak shape, tailing, and low recovery. Although high concentrations of salt and/or organic solvents can minimize these secondary interactions, those conditions may be problematic to the integrity or fidelity of analyzing certain macromolecular analytes4. This application note documents the superior performance of Waters GTxResolve 2000 Å SEC Columns, which are constructed with a large pore size packing material and surface chemistries that minimize nonspecific interactions5. Using Waters DNA Vector Mixture Standard and thyroglobulin protein as representation megadalton macromolecules, the enhanced chromatography performance of GTxResolve 2000 Å SEC 3 µm Columns is demonstrated. The MaxPeak™ High Performance Surfaces (HPS) and novel ethylene bridged polyethylene oxide (HO-PEO) bonded 2000 Å 3 µm particle packing material (Figure 2) of this column technology provide ideal conditions for size-based separations of very large biomolecules, which is now making it possible to establish SEC methods with enhanced efficiency, increased resolution, higher signal-to-noise detection, and improved reproducibility.
Experimental
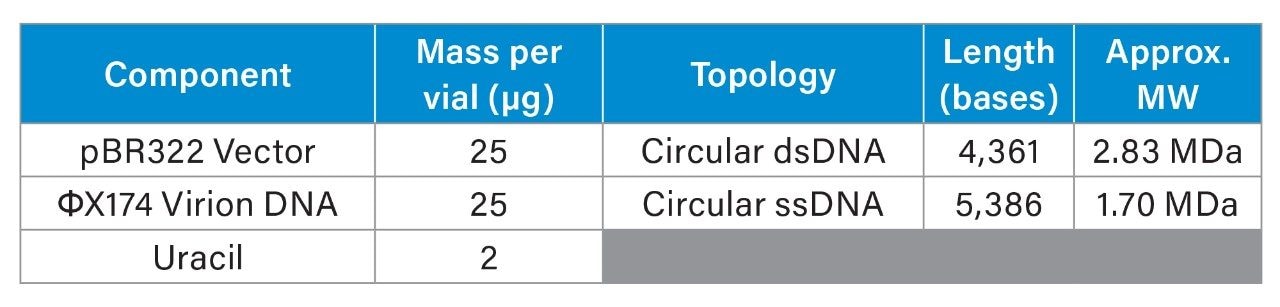
The DNA Vector Mixture Standard (p/n: 186011285) is a 52 µg lyophilized mixture of pBR322 DNA, ΦX174 virion DNA, and uracil (Table 1). pBR322 is a circular double-stranded plasmid DNA (dsDNA) commonly used as a cloning and expression vector in molecular biology. ΦX174 virion DNA is a circular single-stranded DNA (ssDNA) derived from the bacteriophage φX174, often used as a sequencing and molecular weight standard. This standard is suitable to evaluate the GTxResolve 2000 Å SEC MaxPeak Premier 3 µm Columns for chromatographic performance over the megadalton-sized molecular range. The contents of the vial were reconstituted using 100 µL of Milli-Q® water.
LC Conditions
Note on Flow Cell Selection: A 5 mm titanium flow cell (Waters Flowcell, ACQUITY™ PDA, 5 mm, Titanium SKU: 205000613) was used to minimize metal–analyte interactions during SEC-UV analysis in phosphate-buffered mobile phases. Titanium’s chemical inertness reduces the adsorption of nucleic acids and phosphate species, supporting consistent peak shape, signal stability, and reproducible performance, critical for accurate profiling of diverse DNA constructs, including plasmid and viral genomes.
|
LC system: |
ACQUITY Premier System with: – Binary Solvent Manager (BSM, p/n: 186018000 ) – Flow-Through Needle Sample Manager (SM-FTN, p/n: 186018002 ) – ACQUITY UPLC™ Column Manager-A (CM-A, p/n: 186018003 ) |
|
Detector: |
ACQUITY Premier eLambda PDA (p/n: 186018007 ) with 5 mm Titanium Flow Cell (p/n: 205000613) |
|
Column(s): |
– GTxResolve 2000 Å SEC Column, 3 µm, 4.6 x 150 mm (p/n: 176006047) – Agilent® Bio SEC-5 2000 Å, 5 µm, 4.6 x 150 mm |
|
Mobile phase A: |
1× DPBS (HyClone™ Dulbecco's Phosphate Buffered Saline, no calcium and no magnesium, 0.1 µm filtered) |
|
Vials: |
TruView™ pH Control LCMS Certified Clear Glass Vials, 12 x 32 mm, Screw Neck (Waters, p/n: 186005663CV) |
|
Column temperature: |
35 °C |
|
Sample temperature: |
5±3 °C |
|
Flow rate: |
0.1 mL/min |
|
Seal wash: |
10% HPLC-grade methanol / 90% 18.2 MΩ*cm resistivity (Milli-Q®) water (v/v) |
|
Samples and injection volume: |
DNA Vector Mixture (Waters, p/n: 186011285) |
|
Gradient: |
Isocratic |
|
System control and data acquisition: |
Empower™ 3.8.x (tested with 3.8.0.2) |
|
Detection wavelength: |
260 nm |
|
Sampling rate: |
20 points/sec |
Results and Discussion
Improved Resolution
The chromatographic performance of Waters GTxResolve 2000 Å SEC Columns and Agilent Bio SEC-5 2000 Å columns was compared using the Waters DNA Vector Mixture and a simple 1x strength phosphate buffered saline mobile phase. To rigorously compare these column technologies, two manufacturing batches of Waters GTxResolve Columns were tested, each represented by two individual columns. Similarly, a set of two Agilent columns was tested (Figure 3). The Waters GTxResolve SEC Columns demonstrated better performance for key analyte pairs across all tested columns. Specifically, for the resolution between pBR322 aggregates and monomer pBR322 plasmid DNA, the Waters columns provided an average USP resolution of 1.79, outperforming the comparison columns' average resolution of 1.20 by approximately 49%. Similarly, Waters columns exhibited an average USP resolution of 5.93 between the main peaks of pBR322 and φX174 virion DNA, surpassing the comparison column's average of 3.95 by approximately 50% (Figure 3).
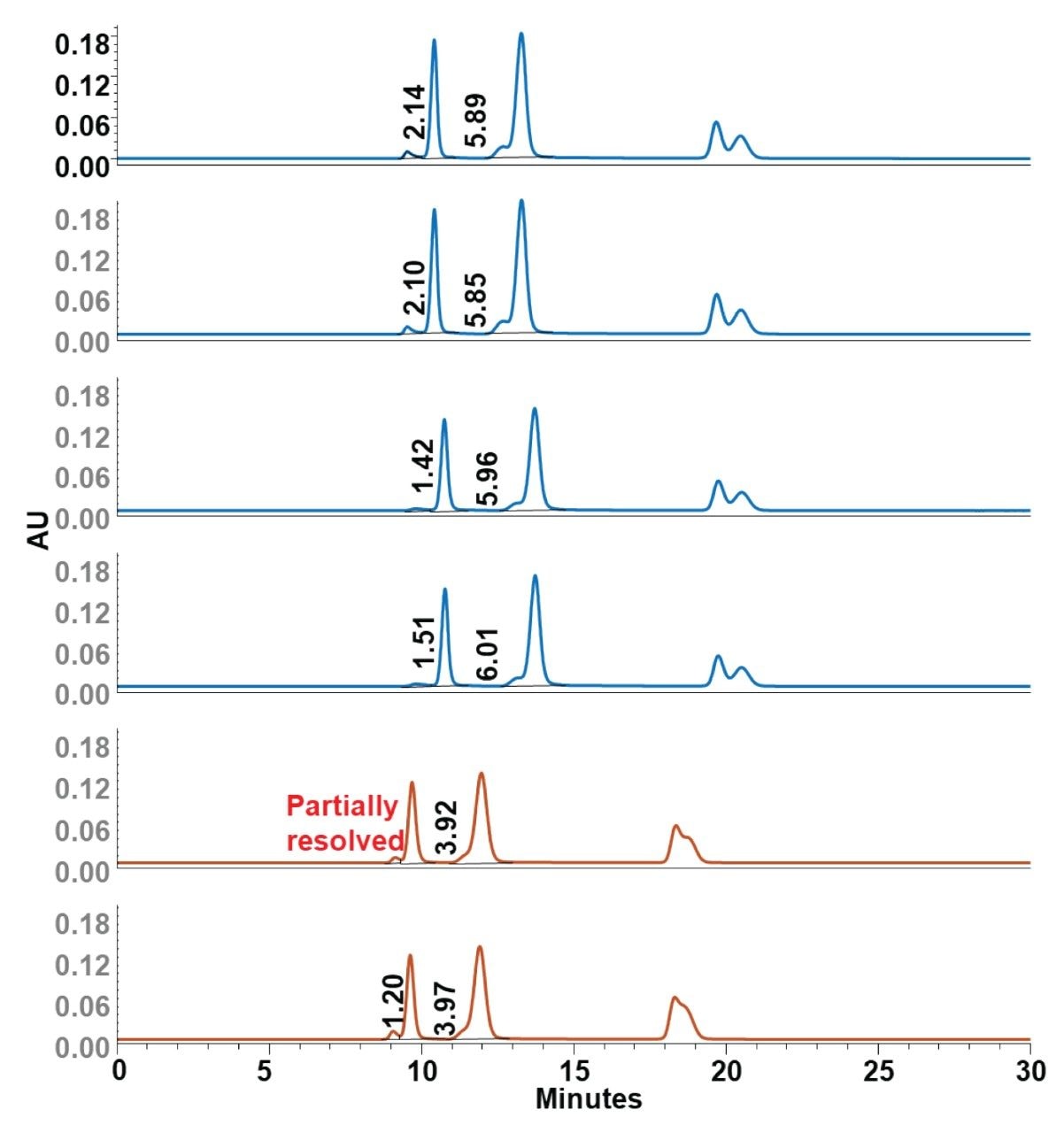
Furthermore, chromatograms obtained from both batches of Waters columns consistently displayed sharp, symmetrical peaks with robust baseline separation, confirming minimal secondary interactions between different batches. In contrast, the comparison columns exhibited broader, less symmetrical peaks and partial resolution of sample components.
Reproducible Column-to-Column Separations
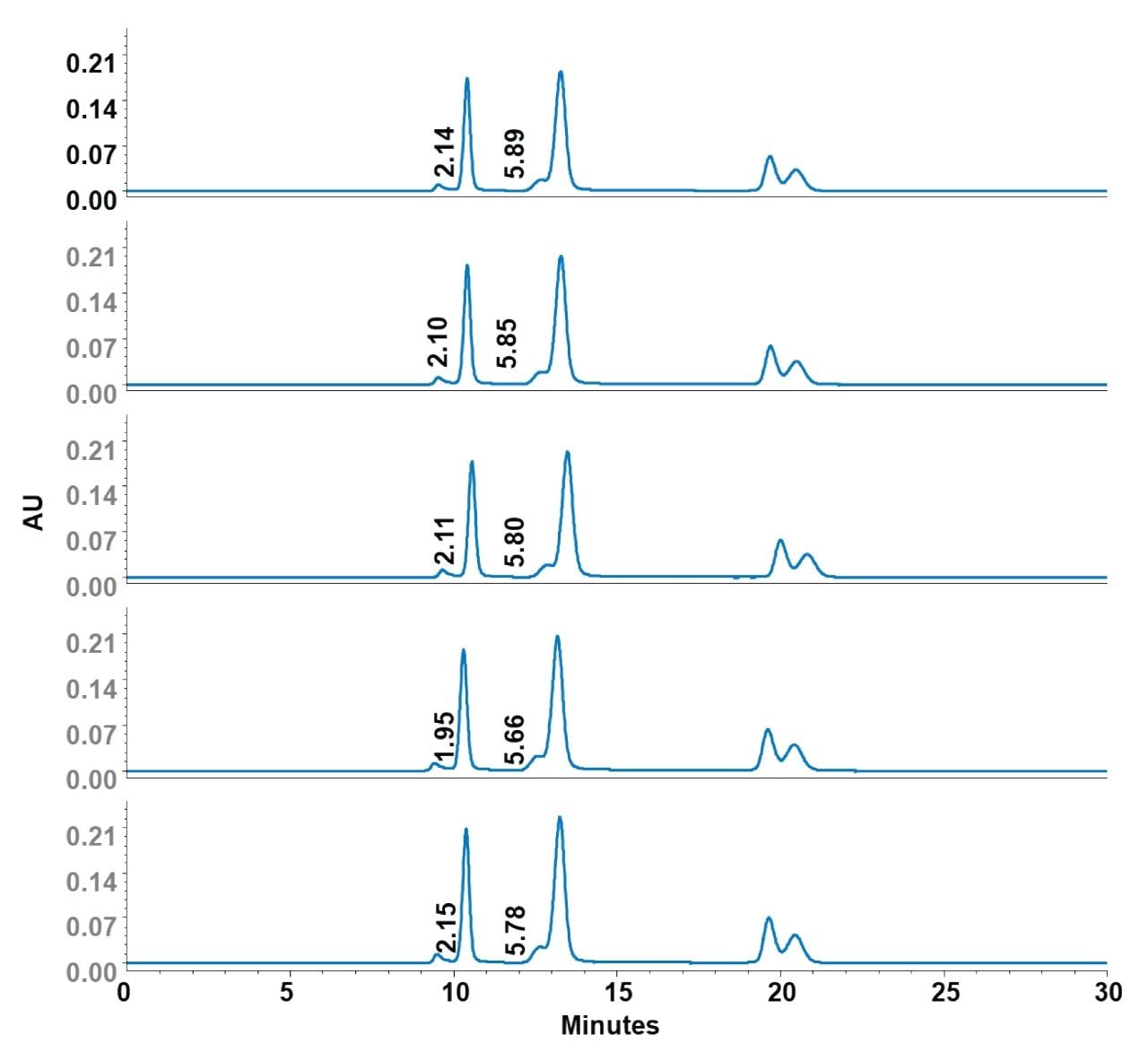
Robust analytical methods require consistent performance across multiple columns. To assess column-to-column reproducibility, separations of the DNA Vector Mixture (pBR322 plasmid DNA, its aggregates, and φX174 virion DNA) were evaluated using five different Waters GTxResolve 2000Å SEC Columns packed from a single batch of packing material. Retention times (RT), relative retention times (RRT), and USP resolutions (HH) were evaluated across columns. The Waters columns displayed exceptional reproducibility, demonstrated by very low percent relative standard deviations (%RSD) in retention times and resolution metrics:
- pBR322 plasmid peak retention time: average RT ~10.37 min, %RSD = 0.89%
- φX174 virion DNA peak retention time: average RT ~13.27 min, %RSD = 0.85%
- Resolution between peaks: consistently achieved USP resolutions around 2.1 for plasmid aggregates/plasmid peak pair and 5.8–5.9 for the plasmid/viral DNA peak pair across columns
Batch-to-Batch Reproducibility
To evaluate manufacturing consistency and batch-to-batch robustness, three Waters GTxResolve 2000 Å SEC Columns prepared with three distinct batches of packing material were tested using the same mixture. Each column was assessed for key chromatographic metrics, including RT, HH, and peak area. Despite being from separate manufacturing batches, all three columns exhibited consistent separation profiles, as evidenced by the following %RSD values:
- pBR322 RT %RSD: 2.83%
- φX174 virion RT %RSD: 3.18%
Chromatographic Performance for Thyroglobulin Monomer / Dimer Separations
Column-to-Column Separations
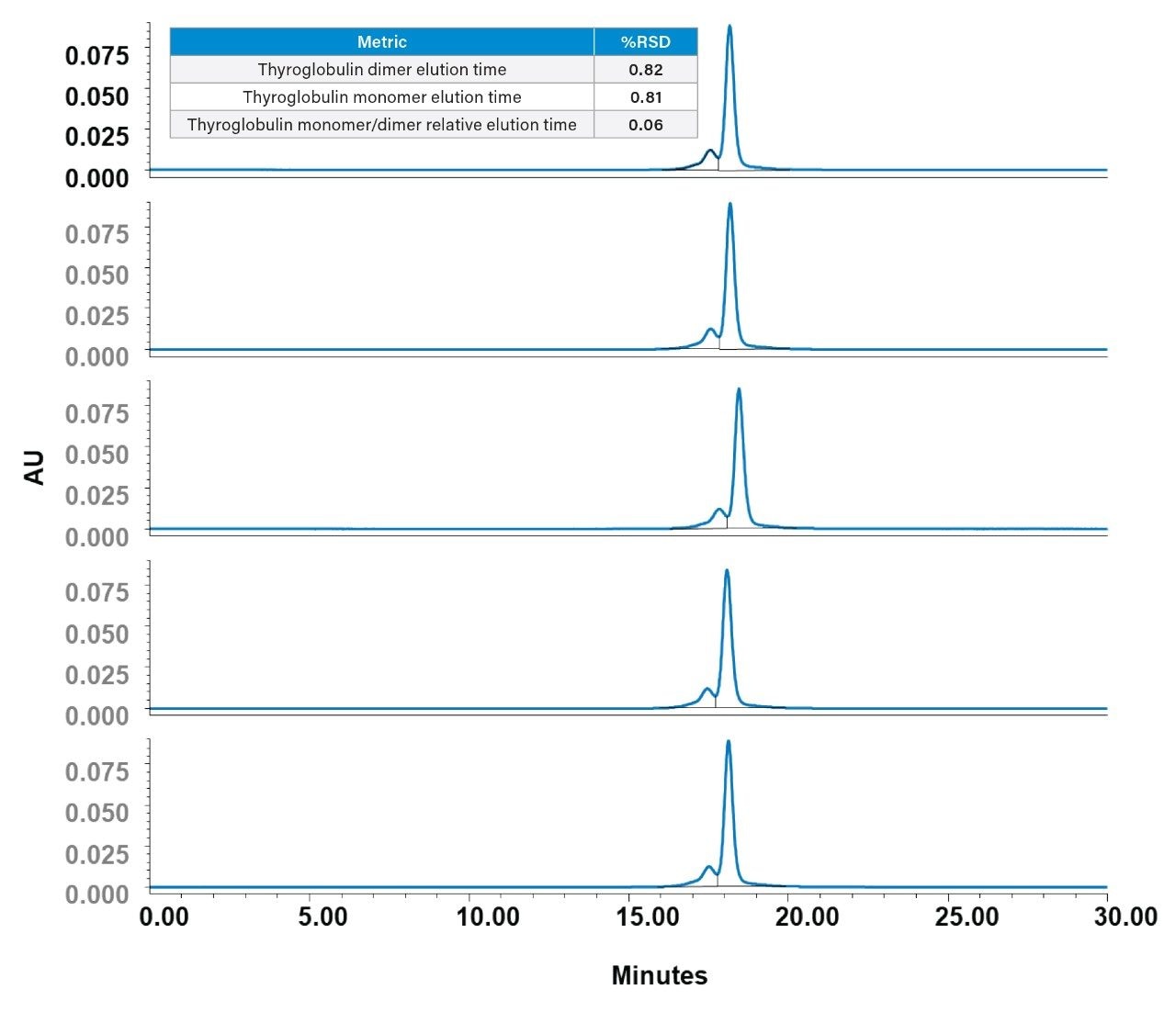
To assess column precision and ensure analytical robustness, thyroglobulin was analyzed across five Waters GTxResolve 2000 Å SEC Columns from a single batch. The results demonstrated exceptional consistency in RT, RRT, and peak area across columns as shown in Figure 5. Superimposing these chromatograms further confirmed the tight alignment of both monomer and dimer peaks, with no shifts in elution profiles and consistent peak symmetry. Plate counts were also consistent, supporting stable column efficiency across multiple columns.
Batch-to-Batch Reproducibility
To further evaluate the performance robustness of Waters GTxResolve 2000 Å SEC Columns across manufacturing batches, thyroglobulin was analyzed on three columns from three different batches. As a high-molecular-weight protein (~669 kDa), thyroglobulin serves as a relevant surrogate for proteinaceous CGT components and biotherapeutics. The presence of both monomeric and dimeric species allows for detailed resolution and reproducibility assessments.
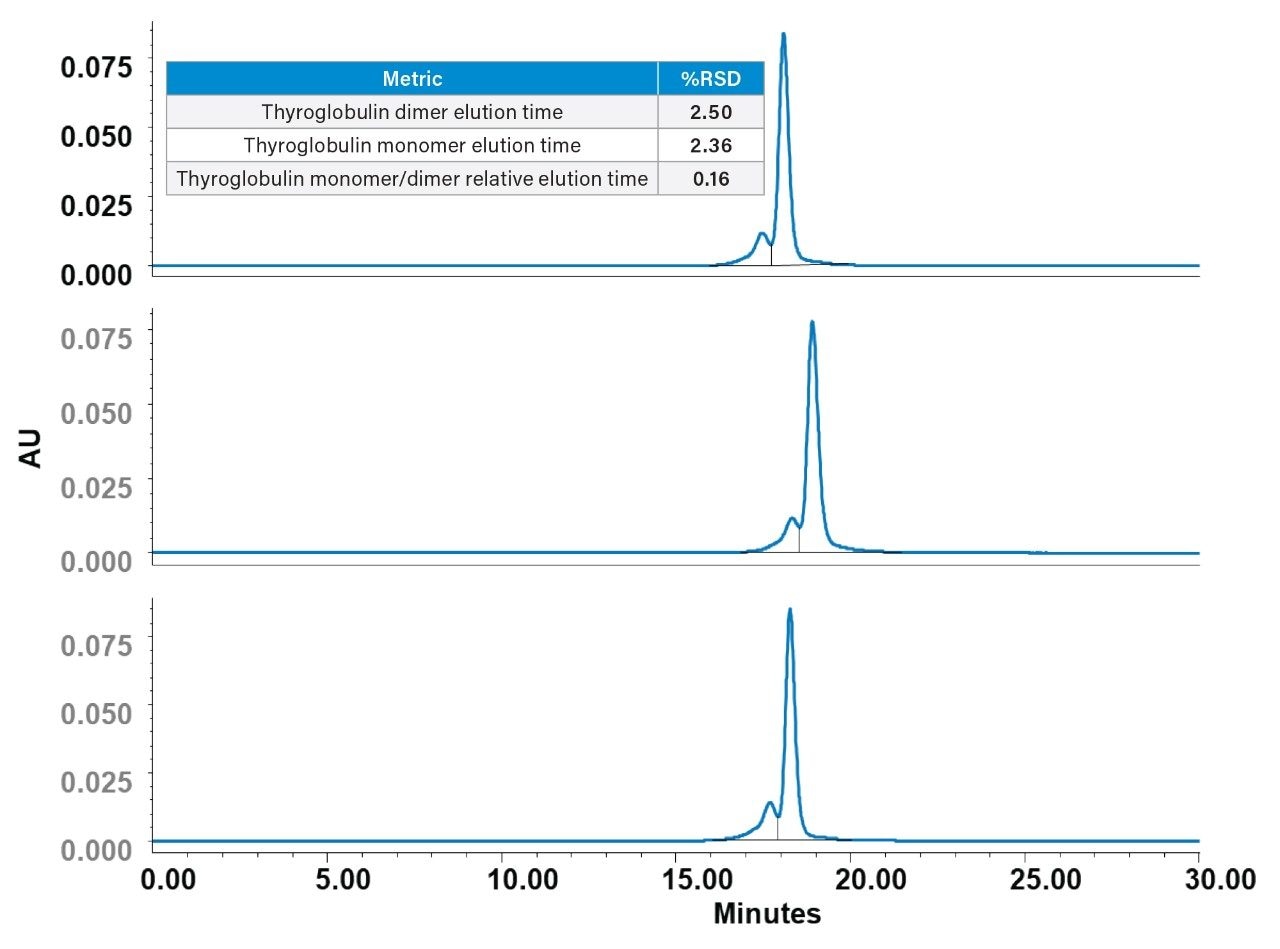
Each column successfully shows the monomer and dimer species of thyroglobulin with excellent consistency in profile and resolution. Quantitative results are summarized in the table in Figure 6.
Out-of-the-Box Performance
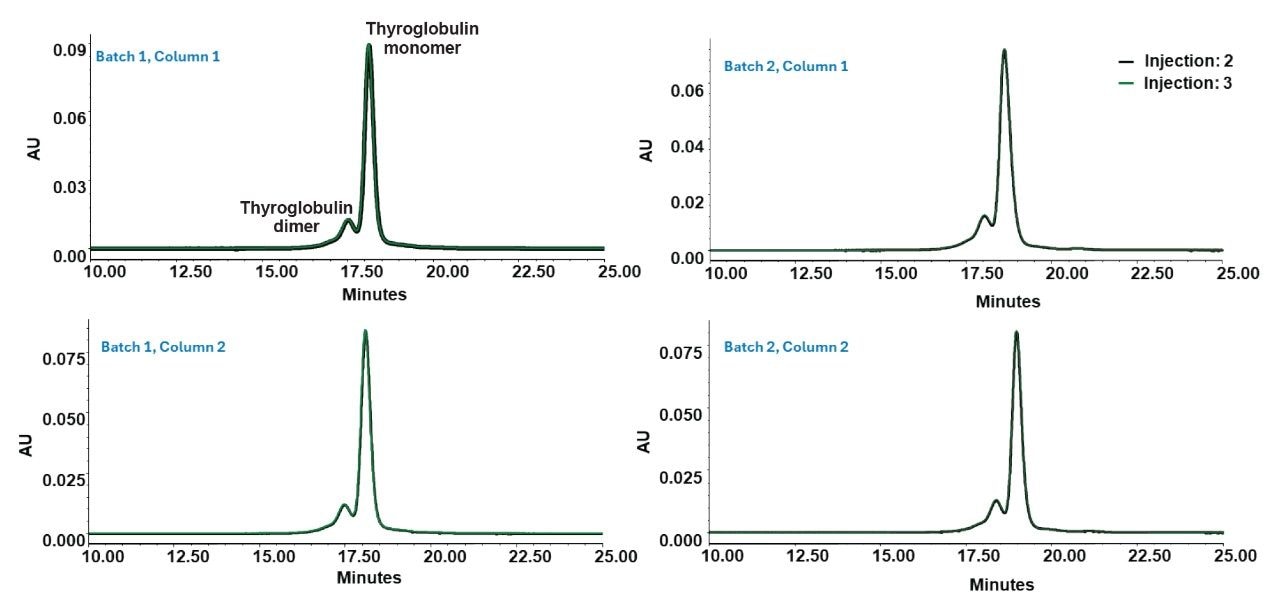
To test the readiness of the GTxResolve 2000 Å SEC 3 µm Columns for immediate analytical use, the percent change in key chromatographic metrics was examined—specifically, the area and RRT between the second and third injections of thyroglobulin monomer and dimer across two column lots from two manufacturing batches out of the box without prior conditioning with biomolecules.
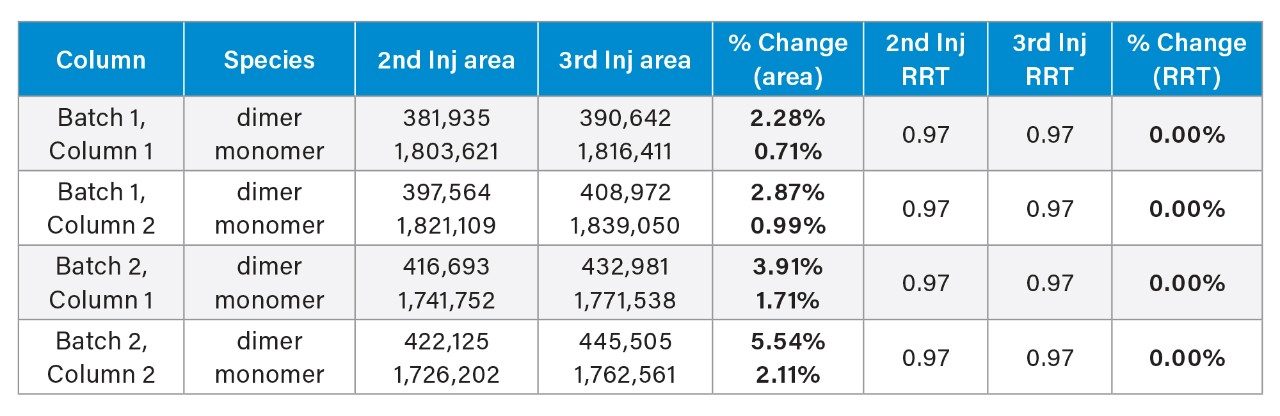
Across all four columns (Batch 1, Columns 1 and 2; Batch 2, Columns 1 and 2), the RRT remained consistent at 0.97 for both dimer and monomer. There wasn’t a significant difference in peak areas between injections 2 and 3. Dimer peak area changes ranged from 2.28% to 5.54%, while monomer peak area changes ranged from 0.71% to 2.11%, as summarized in Table 2. These observations further confirm that the columns deliver reproducible performance without the need for extended conditioning out of the box.
Conclusion
This application note demonstrates the utility of Waters GTxResolve 2000 Å SEC Columns for high-resolution, reproducible analyses of large biomolecular structures relevant to CGT workflows. Using structurally distinct DNA vectors (pBR322 plasmid DNA and φX174 virion DNA), we have confirmed that these GTxResolve Columns facilitate separations with high resolution, minimal secondary interactions, and remarkable reproducibility. Comparative evaluations against other commercially available columns showed that GTxResolve Columns provided significantly improved USP resolution, delivering 20 to 50% greater separation efficiency across critical peak pairs while maintaining stable retention times and consistent chromatographic profiles. In addition to superior performance for nucleic acids, the analysis of thyroglobulin, a high-molecular-weight protein standard, confirmed the column’s robustness across diverse macromolecule types.
Both column-to-column and batch-to-batch reproducibility assessments demonstrated low %RSD values for retention times, relative retention times, and resolution. Furthermore, out-of-the-box performance evaluation showed that back-to-back injections on the columns were highly reproducible and did not require significant conditioning before usage. Additionally, out-of-the-box performance testing revealed that the columns delivered consistent results across sequential injections without requiring extensive conditioning, highlighting their reliability and ease of use. As biopharmaceutical pipelines continue to evolve toward larger and more structurally diverse modalities, the Waters GTxResolve 2000 Å SEC MaxPeak Premier 3 µm Column emerges as a high-performance tool that enables confident characterization of complex macromolecules, supporting critical analytical needs across discovery, development, and quality control environments.
References
- M. Youssef, C. Hitti, J. Puppin Chaves Fulber, and A. A. Kamen. Enabling mRNA Therapeutics: Current Landscape and Challenges in Manufacturing. Biomol. 2023, Vol. 13, Page 1497, vol. 13, no. 10, p. 1497, Oct. 2023, doi: 10.3390/BIOM13101497.
- T. Li et al. CRISPR/Cas9 therapeutics: progress and prospects. Signal Transduct. Target. Ther. 2023 81, vol. 8, no. 1, pp. 1–23, Jan. 2023, doi: 10.1038/s41392-023-01309-7.
- C. Holladay, M. Kulkarni, W. Minor, and A. Pandit. Gene Therapy in Molecular, Cellular, and Tissue Engineering. Vol. 381, no. 5. Massachusetts Medical Society, 2018, pp. 56-1-56–35. doi: 10.1056/NEJMRA1706910.
- A. S. Finny, L. Kizekai, C. Reidy, M. Fasth, B. Addepalli, and M. Lauber. Efficient Profiling of Lipid Nanoparticle Formulations Using Waters GTxResolve 2000 Å SEC Column, MaxPeak Premier 3 µm. Waters Application Note. 720008797. Accessed: May 13, 2025.
- K. J. Camacho et al. Bridged Ethylene Polyethylene Oxide Surfaces to Improve Packing Materials for Widepore Size Exclusion Chromatography. J. Sep. Sci., vol. 47, no. 20, p. e202400541, Oct. 2024, doi: 10.1002/jssc.202400541.
Featured Products
720008847, June 2025