For research use only. Not for use in diagnostic procedures.
This application note describes the sensitive and selective quantification of adalimumab from serum/plasma using a kit-based approach for sample digestion and peptide level clean-up. Coupling different enrichment techniques to the standardized approach of ProteinWorks eXpress Digest and SPE Clean-up Kits provided substantial sensitivity benefit with the increased complexity of each technique.
Using a kit-based approach for the bioanalytical method development of adalimumab simplified and standardized the workflow and facilitated the accurate and robust quantification of adalimumab across multiple, complex sample preparation techniques. This approach enables traditional “small molecule” scientists to quickly and successfully develop LC-MS methods for protein quantification from complex biological matrices.
Adalimumab (HUMIRA) is a humanized monoclonal antibody (mAb) that targets tumor necrosis factor-alpha (TNF-α) and is used in the treatment of inflammation diseases such as rheumatoid arthritis (RA), psoriasis, and Crohn’s disease.1 One of the top selling drugs in the world,2 several of adalimumab’s patents are due to expire in 2017.3 With patent expiries of adalimumab and other popular protein therapeutics nearing, bioanalytical protein quantification has become essential in support of drug research for biosimilar developers and innovator pharma.4-6 In addition, there is high interest to accurately measure mAb concentrations in clinical research as a strategy to optimize the therapeutic dose to improve efficacy or minimize side effects.7,8
Although adalimumab can be quantified with high sensitivity using immunoaffinity (IA) methods, such as ELISA, these assays often suffer from poor standardization and reagent reproducibility across vendors, cross-reactivity, and can take months to years to develop. With multiplexing capability and high specificity, LC-MS is an attractive alternative for the quantification of biologics. The most common strategy to prepare proteins for quantitative MS analysis is the surrogate peptide or bottom-up approach, employing enzymatic digestion and subsequent analysis of the resulting peptides. While widely adopted, this approach can be complex and laborious, requiring optimization of multiple sample processing steps and therefore proves challenging for a traditional “small molecule” scientist to develop sensitive and robust methods. Adding to this complexity, enrichment techniques such as peptide level SPE clean-up and protein level immunoaffinity are often required to achieve ultimate sensitivity or improve robustness. A generic, yet standardized approach to quantify mAbs, amenable to various levels of sample preparation, would greatly reduce method development complexity and facilitate the development of a broadly applicable LC-MS/MS method that could support studies in drug research and development. This application note describes the sensitive and selective quantification of adalimumab from serum/plasma using a kit-based approach for sample digestion and peptide level clean-up. This kitted approach, which can easily be coupled to up-front immunoaffinity enrichment when greater sensitivity is required, was used to achieve adalimumab LLOQs between 5 and 100 ng/mL in serum/plasma.
Preparation of samples, calibration standards and QC samples
Calibration curve standards and quality control (QC) samples of adalimumab were prepared at various concentration levels (1–500,000 ng/mL) in rat plasma and human serum. A stable isotope labeled (15N13C) monoclonal antibody, SILu MAb, was used as the internal standard (ISTD). All calibration curve standards, QC levels, and blank (non-spiked) serum/plasma samples were prepared in triplicate. Three sample preparation strategies were employed: (1) Direct digestion (no protein level clean-up) of human serum and subsequent peptide level SPE sample enrichment, (2) generic protein level immunoaffinity enrichment (Protein A) in human serum followed by digestion and peptide level SPE enrichment, and (3) specific protein level immunoaffinity enrichment (anti-hIgG FC capture) in rat plasma followed by digestion. These workflows are described in greater detail below.
Sample preparation method 1: Direct digestion + SPE
Adalimumab spiked human serum samples (75 μL) were digested using the ProteinWorks eXpress Direct Digest Kit (p/n 176003688) and included 5-step protocol. Post-digestion purification of signature peptides was done using the ProteinWorks μElution SPE Clean-Up Kit (p/n 186008304). Specifically, 50 μL of the post-digestion sample was processed by SPE, eluted with 50 μL of elution solution, and diluted with 50 μL of water prior to LC-MS analysis.
Sample preparation method 2: Generic protein level clean-up + digestion + SPE
Adalimumab was immunopurified from human serum (75 μL) using a 96-well Protein A agarose-based plate (GE Healthcare p/n: 28-9031-33) using the vendor supplied protocol. 300 μL of the post-affinity purified sample was neutralized (pH 8) and evaporated by vacufuge. The post-affinity purified serum was then digested using the ProteinWorks eXpress Digest Kit (p/n 176003689). 120 μL of digestion buffer was used to reconstitute the immunopurified sample which was subsequently digested using the supplied 5-step ProteinWorks protocol. Post-digestion purification of signature peptides was done using the ProteinWorks μElution SPE Clean-up Kit and included protocol. Specifically, 140 μL of the post-digestion sample was processed by SPE, eluted with 50 μL of elution solution, and diluted with 50 μL of water. Resulting samples were then injected for LC-MS analysis.
Sample preparation method 3: Specific protein level clean-up + digestion
Adalimumab was immunopurified from rat plasma (100 μL) using a goat derived anti-human biotinylated IgG antibody conjugated to a streptavidin bead slurry (Promega p/n V7820 and V7830). Following affinity purification, samples were digested using ProteinWorks eXpress Digest Kit and included 5-step protocol. Specifically, 50 μL of affinity purified sample was neutralized (pH 8), diluted to 120 μL with ProteinWorks digestion buffer, and subsequently digested using the supplied 5-step protocol. Resulting samples were then injected for LC-MS analysis.
|
LC system: |
ACQUITY UPLC |
|
Detection: |
Xevo TQ-XS Mass Spectrometer, ESI+ |
|
Column: |
ACQUITY UPLC Peptide BEH C18, 300Å, 1.7 μm, 2.1 x 150 mm |
|
Column temp.: |
55 °C |
|
Sample temp.: |
15 °C |
|
Injection vol.: |
10 μL |
|
Mobile phases: |
A: 0.1% Formic acid in H2O, B: 0.1% Formic acid in ACN |
|
Time (min) |
Flow rate (mL/min) |
%A |
%B |
Curve |
|---|---|---|---|---|
|
Initial |
0.3 |
95.0 |
5.0 |
6.0 |
|
1.5 |
0.3 |
95.0 |
5.0 |
6.0 |
|
9.5 |
0.3 |
65.0 |
35.0 |
6.0 |
|
10.0 |
0.3 |
10.0 |
90.0 |
6.0 |
|
11.0 |
0.3 |
10.0 |
90.0 |
6.0 |
|
11.5 |
0.3 |
95.0 |
5.0 |
6.0 |
|
13.5 |
0.3 |
95.0 |
5.0 |
6.0 |
|
MS system: |
Xevo TQ-XS |
|
Ionization mode: |
ESI+ |
|
Capillary: |
2.9 kV |
|
Cone: |
32 V |
|
Source offset: |
30 V |
|
Source temp.: |
150 °C |
|
Desolvation temp.: |
600 °C |
|
Cone gas flow: |
150 L/hr |
|
Desolvation gas flow: |
1000 L/hr |
|
Collision gas flow: |
0.15 mL/min |
|
Nebulizer gas flow: |
7 Bar |
|
Data management: |
MassLynx (v4.1) |
|
Quantification software: |
TargetLynx |
With impending US patent expiry of adalimumab in 2017, the focus on this drug in pharma, CROs, biosimilar research, and clinical research labs has increased. Adalimumab’s pharmacokinetics is characterized by a rapid distribution, low clearance, long half-life (~2 weeks), and limited tissue distribution9 with serum trough levels reported over a large dynamic range (20 to ~10,000 ng/mL).9-11 There is a strong need to quickly develop methods which can sensitively and accurately measure adalimumab in biological matrix across a broad dynamic range to support drug discovery and clinical research activities.
While LC-MS is widely adopted for small molecule and peptide bioanalytical quantification, its use for protein quantification remains limited due to the challenges associated with the complex and laborious sample preparation workflows. Additionally, achieving accurate and robust methods which reach the desired sensitivity limits in biological matrices can be difficult. To date, there is no universal or standardized sample preparation and LC-MS workflow amenable to the diversity of protein therapeutics, making method development particularly challenging. In this application note, we highlight the benefits of using sample enrichment with protein level immunoaffinity and a kit-based approach with ProteinWorks eXpress Digest and μElution SPE Clean-up Kits for the quantification of adalimumab from serum/plasma.
Identification and optimization of signature tryptic peptides derived from the protein of interest are a critical aspect needed for successful MS method development. This process can be complex and challenging, particularly for a traditional “small molecule” bioanalytical scientist. To simplify this process, Skyline (MacCoss Labs, University of Washington),12 an open-access software, was used to predict and develop an LC-MS method for adalimumab. Using Skyline, an in-silico tryptic digestion of adalimumab was performed to predict: tryptic peptide sequences, charge states, fragment ions, and collision energies for MRM transitions. The resulting tryptic peptide sequences were then compared to the human plasma proteome (NCBI BLAST)13 to exclude peptides which were not unique to adalimumab (present in the plasma proteome). The full amino acid sequence of adalimumab1 and its unique signature peptides: APYTFGQGTK, NYLAWYQQKPGK, GLEWVSAITWNSGHIDYADSVEGR (highlighted in blue), are illustrated in Figure 1. Following unique signature peptide identification, development and optimization of the MRM method was experimentally determined using a Skyline/MassLynx workflow performed on a Xevo TQ-XS Tandem Quadrupole MS using a tryptic digest of adalimumab in buffer and in plasma matrix. Adalimumab tryptic peptides used for quantification were ultimately chosen on the basis of their signal intensity, selectivity in matrix, and chromatographic performance. Because it is a fully humanized mAb, adalimumab is particularly difficult to quantify in human serum/plasma, as it shares very close sequence homology with endogenous human IgG. This severely restricts the tryptic peptide options available for adalimumab’s quantification. Ultimately, three unique tryptic peptides of adalimumab were chosen for its quantification. Optimized MS conditions and MRM transitions for the adalimumab and SILuMAb (ISTD) tryptic peptides are listed in Table 1. MRM transitions used for quantification are highlighted in blue.

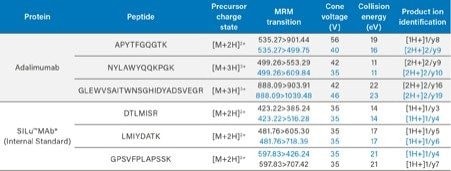
Table 1. Final MS conditions for adalimumab and SILuMAb tryptic peptides, including precursor and fragment ions. MRM transitions used for quantification are highlighted in blue.
*15N13C stable isotope labeled lysines and arginines.
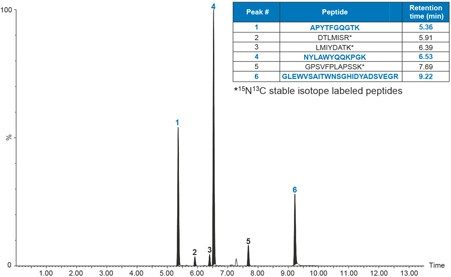
For the present method, the goal was to develop an LC method which could achieve the analytical sensitivity desired, chromatographically resolve endogenous interferences, and maintain adequate throughput. Chromatographic separation of adalimumab and SILuMAb tryptic peptides was achieved using an ACQUITY UPLC Peptide BEH C18, 300Å, 1.7 μm, 2.1 x 150 mm Column. Figure 2 highlights the chromatographic separation of the three adalimumab (highlighted in blue) and three SILuMAb peptides. A long, shallow gradient from 5 to 35% B over 8 minutes afforded the best chromatographic performance possible for all three adalimumab peptides, each with peak widths <5 seconds wide. With so few unique signature peptides for adalimumab, it was important to ensure that all peptides were quantified reproducibly, in some cases at the expense of overall sensitivity. For example, quantification of the top performing peptide, GLEWVSAITWNSGHIDYADSVEGR, could be improved by increasing the percentage of organic at the start of the gradient and making the gradient shallower. However, this severely impacted the chromatographic performance and reproducibility of the APYTFGQGTK and NYLAWYQQKPGK peptides.
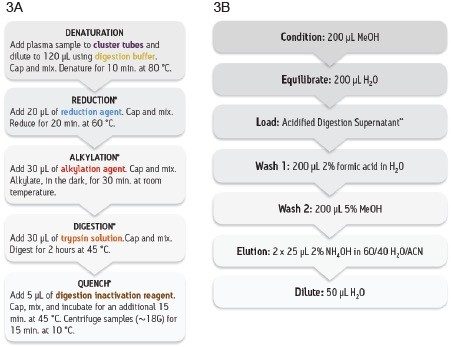
Developing an effective sample preparation strategy for LC-MS protein quantification which is accurate and reproducible can be quite challenging. Due to the high complexity of biological matrices with hundreds of endogenous proteins, the complexity of the protein therapeutic, and the broad range of sensitivity desired (low ng/mL–μg/mL), various sample analytical techniques are often required (e.g., immunoaffinity enrichment, sample digestion, and SPE). With numerous endogenous interferences present in serum/plasma, and the limited unique signature peptides available for adalimumab quantification, three sample preparation methods were assessed for its quantification: (1) Direct digestion (no protein level clean-up) of human serum and subsequent peptide level SPE clean-up, (2) Generic protein level clean-up (Protein A) in human serum followed by digestion and SPE peptide level clean-up, and (3) Specific affinity capture (anti-hIgG FC) in rat plasma followed by digestion. To simplify and standardize the digestion and SPE sample preparation, we have used the ProteinWorks eXpress Digest (5-step protocol) and μElution SPE Clean-up Kits. The ProteinWorks digest and SPE protocols are shown in Figures 3A and 3B, respectively.
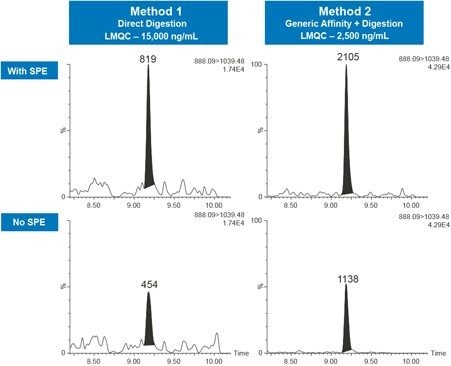
While accurate and reproducible quantification of adalimumab can be achieved with direct digestion and/or generic affinity and subsequent digestion, sensitivity and specificity can be limited. Due to the large protein load in serum/plasma, endogenous interferences are high and compete for signal in the mass spectrometer, leading to lower signal for the peptides of interest. Employing peptide level clean-up with SPE can further decrease sample complexity, remove potential matrix interferences, and provide sample concentration. This benefit is illustrated in Figure 4 for both Method 1 (Direct Digestion) and Method 2 (Generic affinity + Digestion), respectively. Using SPE clean-up, a 2x increase in signal intensity for the GLEWVSAITWNSGHIDYADSVEGR peptide can be seen.
For all three sample preparation methods, accurate, linear, and precise quantification was achieved. Calibration curves from the three adalimumab tryptic peptides prepared using all three methods were linear over a range of 1.0 to 4.0 orders of magnitude with R2 values >0.99 using 1/X2 weighted regressions. Mean accuracies for all standard curves ranged from 88.0–111.1%. A summary of standard curve performance for the APYTFGQGTK, NYLAWYQQKPGK, and GLEWVSAITWNSGHIDYADSVEGR tryptic peptides is listed in Table 2.
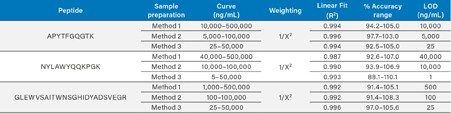
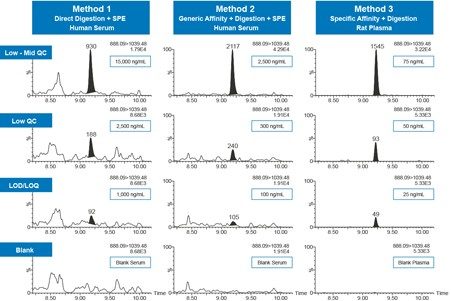
Using the simplest sample preparation approach, with direct digestion and SPE (Method 1), quantification limits from 1,000–40,000 ng/mL were achieved. Employing generic affinity capture, digestion, and SPE (Method 2) afforded quantification limits which were an order of magnitude lower than Method 1, with quantification limits between 100–10,000 ng/mL. Ultimate sensitivity was realized with specific affinity capture and subsequent digestion (Method 3), achieving quantification limits between 5–25 ng/mL.
QC performance for Methods 1–3 is highlighted in Table 3 (panels A–C), while QC chromatographic performance for the GLEWVSAITWNSGHIDYADSVEGR tryptic peptide is highlighted in Figure 5. QC accuracy and precision performance for the three sample preparation methods met FDA and EMA guidelines14,15 with mean % RSDs <15% and % QC accuracy ranges of 91.4–107.0 (Method 1), 91.4–108.3 (Method 2), and 88.0–111.1 (Method 3), respectively.
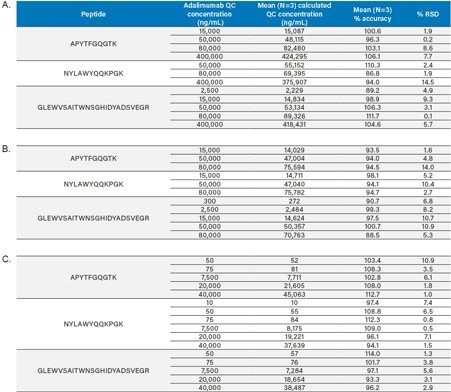
This application assesses the sensitivity gains of three different sample preparation techniques for the quantification of adalimumab in serum/ plasma. Coupling different enrichment techniques to the standardized approach of ProteinWorks eXpress Digest and SPE Clean-up Kits provided substantial sensitivity benefit with the increased complexity of each technique. Using a typical set of standard curve and QC samples in serum/ plasma, limits of quantification between 5–100 ng/mL were achieved when employing an up-front immunoaffinity capture. Across all techniques (Methods 1–3), excellent linearity and precision with % RSDs <15% for all QCs were achieved. Using a kit-based approach for the bioanalytical method development of adalimumab simplified and standardized the workflow and facilitated the accurate and robust quantification of adalimumab across multiple, complex sample preparation techniques. This approach enables traditional “small molecule” scientists to quickly and successfully develop LC-MS methods for protein quantification from complex biological matrices.
720006210, March 2018