This is an Application Brief and does not contain a detailed Experimental section.
This work demonstrates that incorporating ProMass HR in MassLynx Software data processing can be used for synthetic peptide impurity analysis.
High-resolution characterization of synthetic peptide impurities using MassLynx Software and ProMass HR.
The potential for therapeutic peptides as personalized vaccines has been demonstrated with the recent successful adjuvant treatment of melanoma.1 However, impurities introduced through the synthetic process or degradation of the peptide can impact the efficacy and consistency of the drug product, making it critical to characterize and monitor the impurity profiles of synthetic peptides for drug development and quality control. Mass spectrometry (MS) is a commonly used platform in pharmaceutical development for structural characterization, molecular weight confirmation, and impurity analysis. Workflows that are intuitive and can be automated are an effective means to assist the processing and interpretation of complex MS-based data enabling biopharmaceutical companies to bring life-saving drug products to market faster with increased confidence in product quality.
In this study, we used a Waters Xevo G2-XS QTof MS coupled with an ACQUITY UPLC H-Class Bio System and a TUV Detector for high-mass resolution synthetic peptide impurity profiling. The software ProMass HR (by Novatia) for MassLynx Software serves as an automated data processing platform for MS spectral deconvolution, impurity analysis, and reporting.2 A synthetic peptide, eledoisin (purchased from New England Peptide), was selected as a model analyte for its clinical relevance. As shown in Figure 1, with a 20 min gradient at 60° C from 18-28% acetonitrile modified with 0.1% formic acid, the main peak of eledoisin was separated from six impurity peaks. For users with UNIFI Scientific Information System, data can be exported as .raw file and processed using MassLynx in conjunction with ProMass HR.
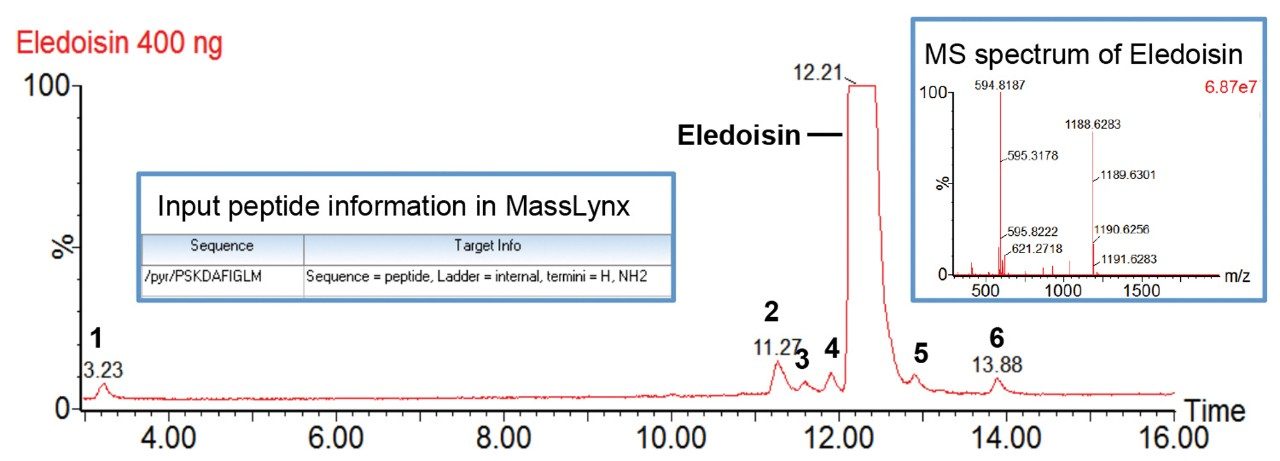
For MassLynx Software, data acquisition and processing can be performed on MassLynx integrated with ProMass HR as described previously.3 Briefly, this process includes inputting target information in MassLynx (Figure 1), setting up the ProMass Bridge Parameter file, and entering unnatural or modified amino acids in the ProMass Parameter file. In addition, to fully utilize the resolving power of a Xevo G2-XS, a universal PPL (positive probability ltd) method for synthetic peptides was created in ProMass HR to enable deisotoping of high resolution mass spectra, noise removal, and charge deconvolution. The impurity profiling results were summarized in a report page generated by ProMass HR (Figure 2). As shown in the target summary table, the target peptide eledoisin was successfully identified in the chromatogram with a mass accuracy of 2.4 ppm. Information such as target peptide confirmation, impurity identification, purity assessment, deconvoluted MS spectra, and peak reports can be accessed via hyperlinks or summarized in a report format demonstrating ProMass HR, when used in conjunction with MassLynx, offers high throughput characterization and monitoring of synthetic peptide impurities.
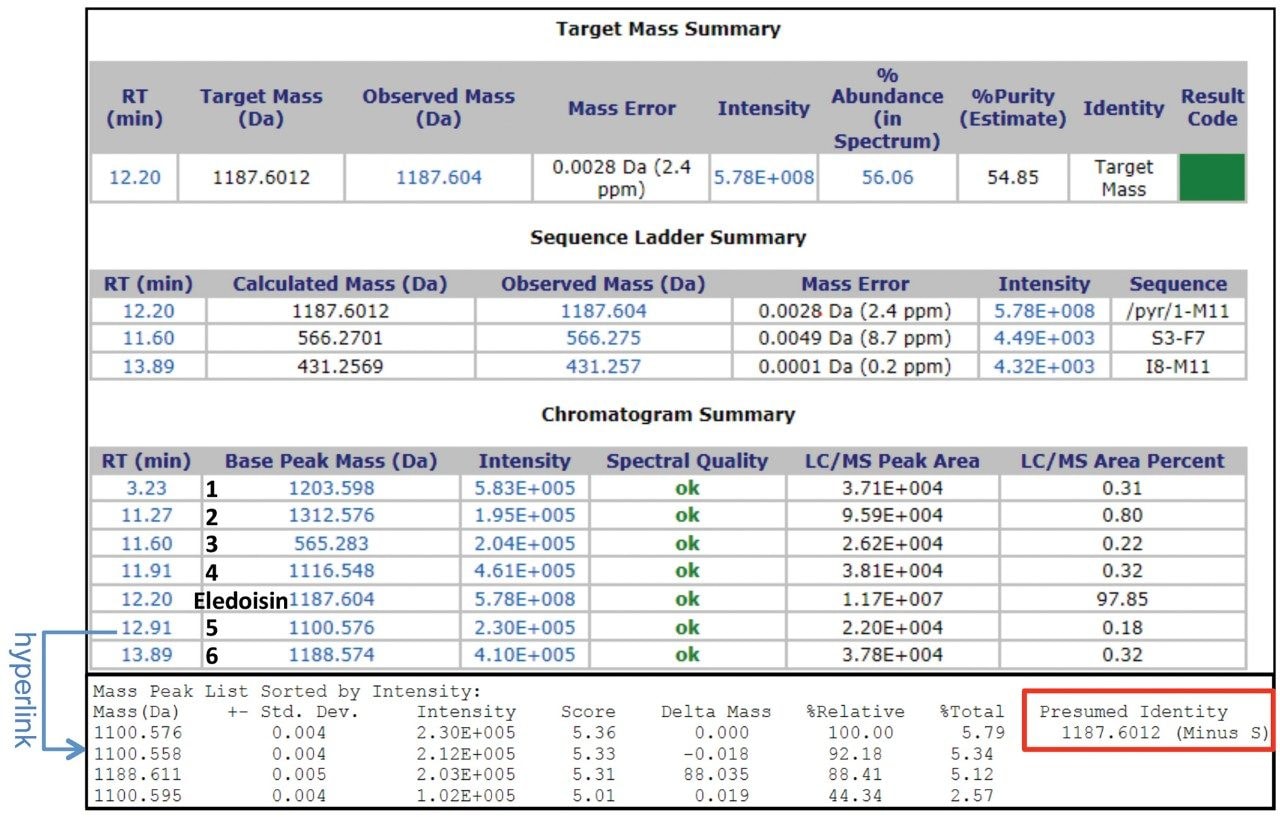
This work demonstrates a high throughput LC-UV/HRMS workflow for synthetic peptide mass confirmation and impurity profiling using a Xevo G2-XS QTof MS with UNIFI or MassLynx and ProMass HR. Retention time, mass, and presumed identity of both the target peptide and impurities were summarized in reports generated by ProMass HR. To this end, the developed workflow can be used for high-resolution synthetic peptide impurity profiling in pharmaceutical developments.
720006104, October 2017