HILIC UPLC/FLR/QTof MS is a powerful glycan characterization tool for characterization of complex glycans samples. Conventional HPLC method lacks the resolution power and sensitivity. Fractionation for MS analysis step is eliminated since the ACQUITY UPLC System is directly interfaced into a Xevo QTof MS. SimGlycan Software is part of the system solution, which interprets the collision fragmentation data, and offers a mean to elucidate glycan structures.
Glycosylation of therapeutic protein drugs is of particular importance because it plays a vital role in the clinical performance of these drugs. In this work, we study the N-linked glycans from two Coagulation Factor IX biologics that are used for Hemophilia B treatment; one is recombinant (rFIX, BeneFIX) and the other one is derived from human plasma (pd-FIX, Mononine). Both Factor IX proteins are heavily glycosylated (Figure 1).1 Previous findings on their glycoforms were done primary using orthogonal HPLC separation techniques, typically via Ion Exchange chromatography (IEX) and hydrophilic interaction (HILIC) modes, due to the complex nature of the Factor IX glycans. For mass profiling and structure characterization, mass spectrometry (MS) was typically used offline for LC fractions.
Waters has developed a HILIC UPLC/FLR/QTof MS analytical platform for fluorescent-labeled glycan characterization. Significant improvements can be made such as peak resolution, speed, sensitivity, and the ability to identify and quantify even the minor glycans. An ACQUITY UPLC System is directly interfaced to a Xevo QTof MS, eliminating the need for fractionation. Comparative analysis of rFIX and pd-FIX glycans using this platform is demonstrated.
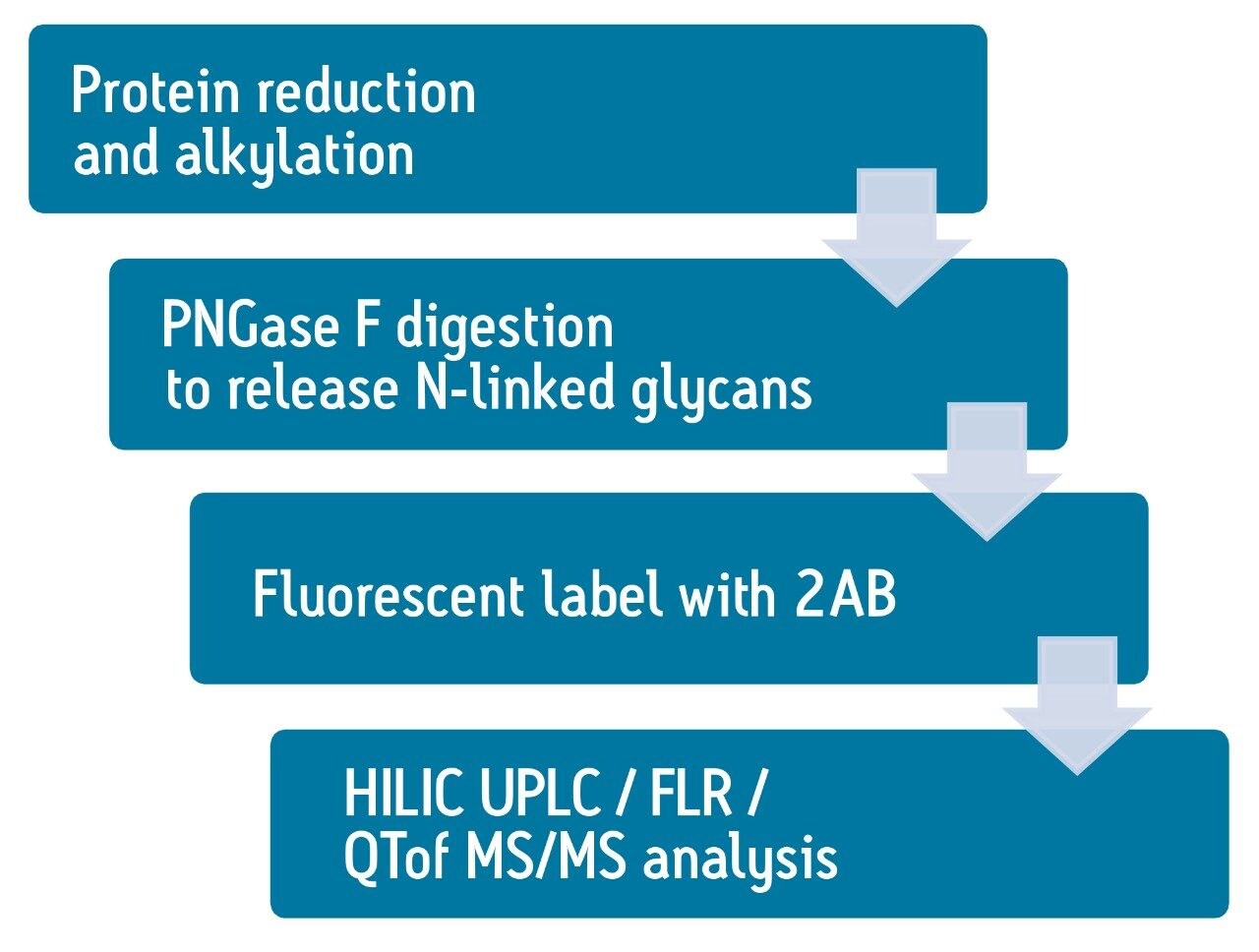
Figure 2 shows the basic glycan analysis workflow. FIX proteins are reduced and alkylated using DTT and IAM, followed by PNGase F enzymatic digestion overnight to release the glycans. The glycans are extracted using HILIC SPE device and labeled with 2-aminobenamide (2AB) dye, and the excess dye was removed by HILIC SPE again.2
|
LC System: |
ACQUITY UPLC |
|
Detection: |
ACQUITY UPLC FLR |
|
Column: |
ACQUITY UPLC BEH Glycan Column (2.1 x 150 mm) |
|
Column temp.: |
60 °C |
|
Sample temp.: |
10 °C |
|
Injection volume: |
5 μL |
|
Flow rate: |
0.4 mL/min |
|
Mobile phase A: |
100 mM Ammonium Formate (~ pH 4.3) |
|
Mobile phase B: |
100% Acetonitrile (Fisher Optima) |
|
Time (min) |
Flow rate |
%A |
%B |
Curve |
|
Initial |
0.400 |
39.0 |
70.0 |
- |
|
60.00 |
0.400 |
50.0 |
50.0 |
6 |
|
60.10 |
0.250 |
95.0 |
5.0 |
6 |
|
63.00 |
0.250 |
95.0 |
5.0 |
6 |
|
64.00 |
0.300 |
30.0 |
70.0 |
6 |
|
65.00 |
0.400 |
30.0 |
70.0 |
6 |
|
72.00 |
0.400 |
30.0 |
70.0 |
6 |
|
MS system: |
Xevo QTOF MS |
|
Ionization mode: |
ESI positive |
|
Capillary voltage: |
3200 V |
|
Cone voltage: |
30 V |
|
Desolvation temp.: |
350 °C |
|
Desolvation gas: |
800 L/Hr |
|
Source temp: |
100 °C |
|
Acquisition range: |
700 to 2000 m/z |
|
Collision energies: |
4 V |
|
Lockmass: |
Cesium Iodide (CSI): 1 μg/μL in 50% MeCN |
MassLynx 4.1
SimGlycan v. 2.9 (Premier Biosoft)
rFIX and pd-FIX has very different glycans profiles (Figure 3, 4). Glycans released from rFIX are mostly fucosylated bi-, tri-, and tetra-antennary complex type glycans. Additional lactosamine units were observed in some glycans. Man5 and a core N-glycans with an addition of fucose were also identified (Table 1). Glycans from pd-FIX are show more heterogeneity, especially for the larger glycans.
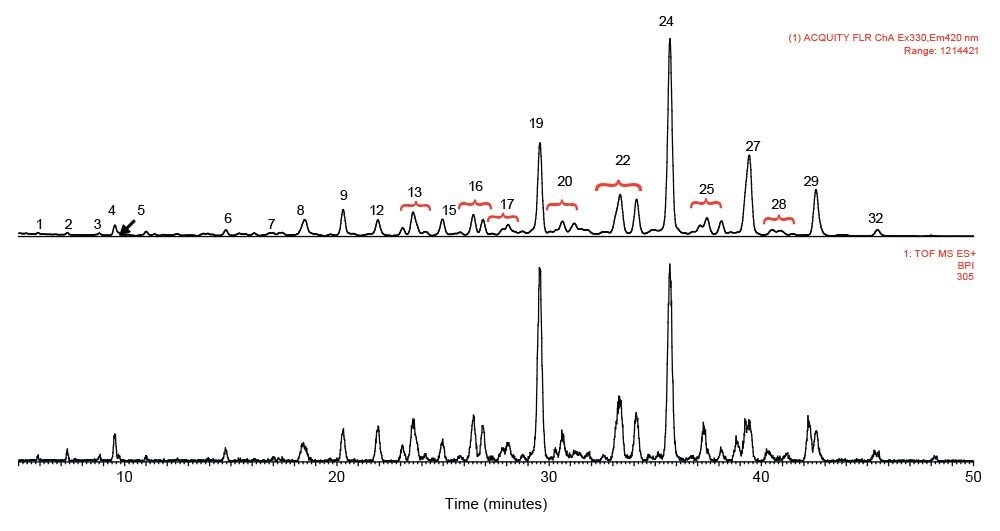
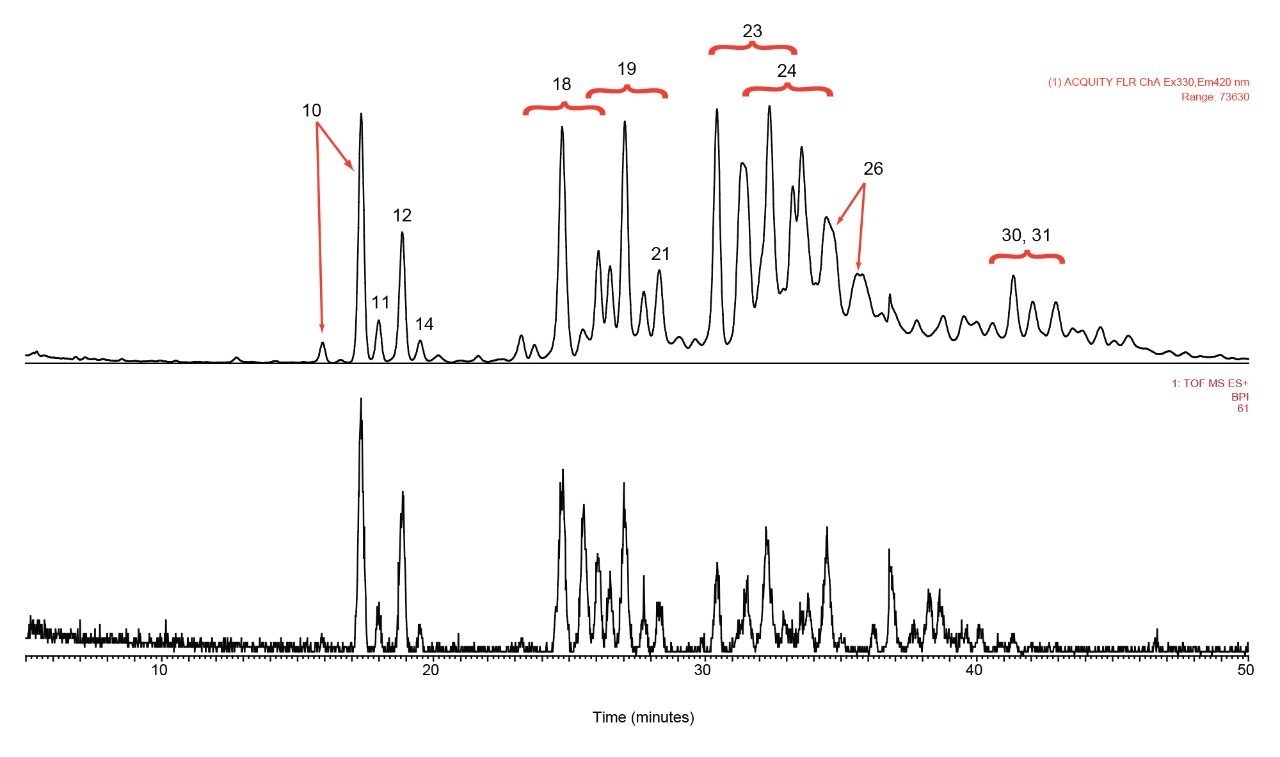
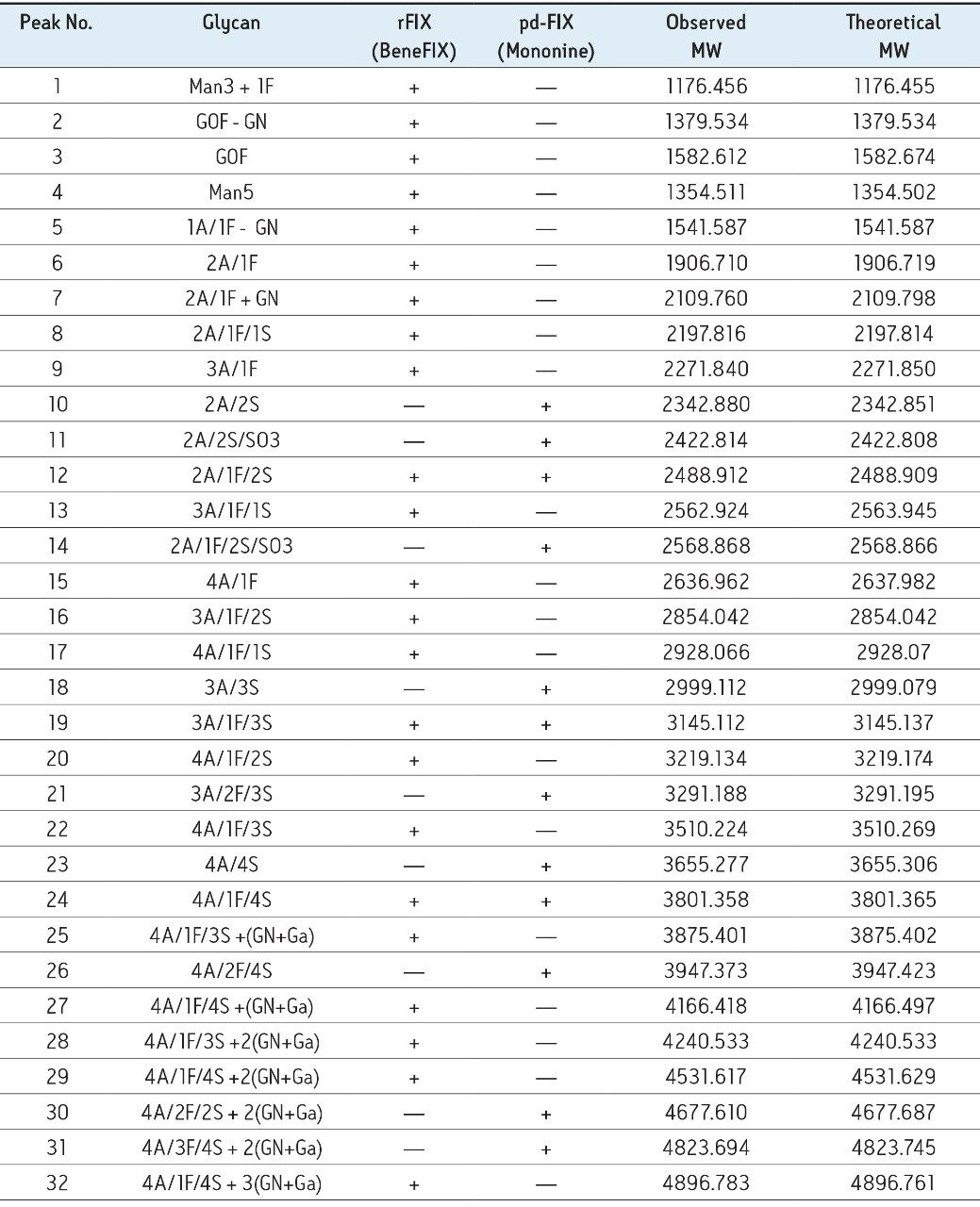
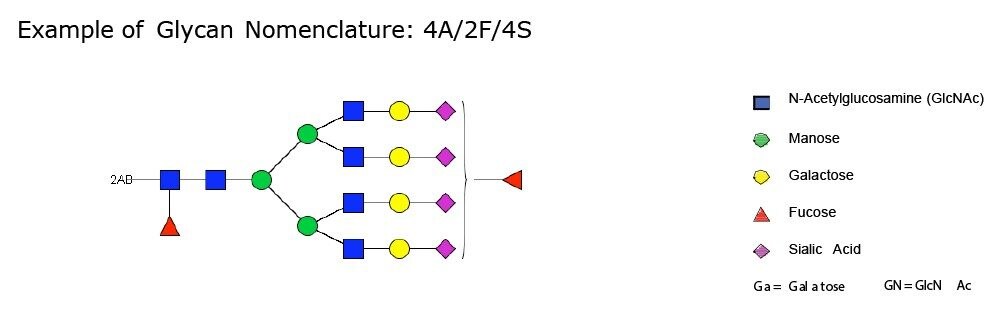
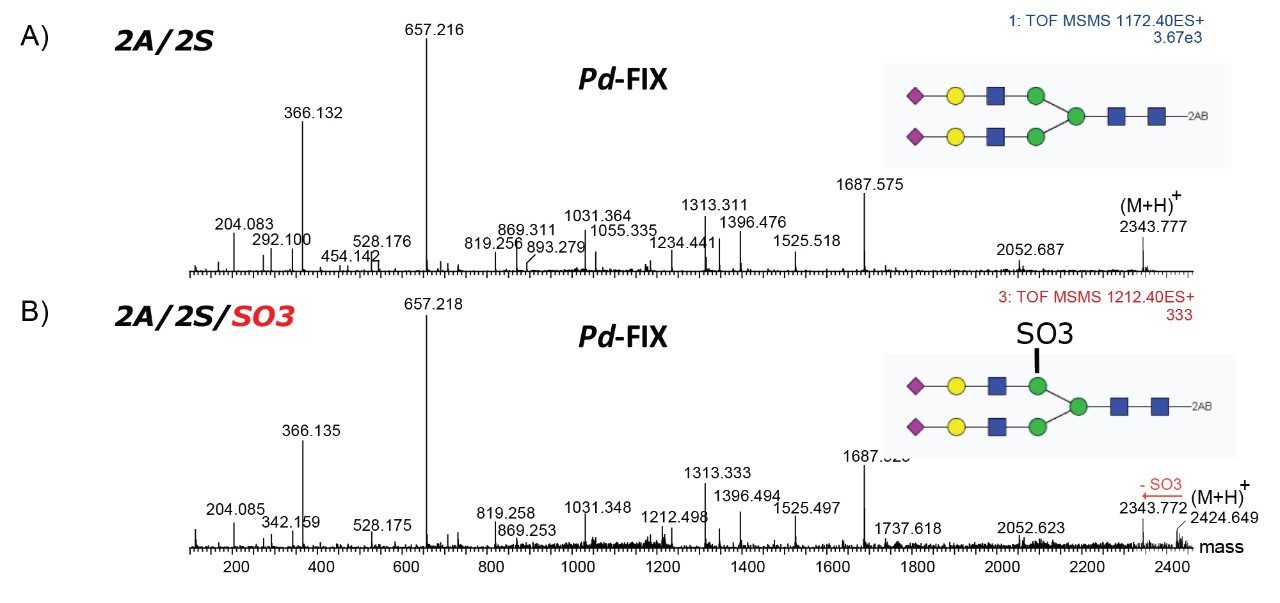
Only the complex type glycans with polylactosamine units were observed from pd-FIX; the level of fucosylation is from 0 to 2 (Table 1). Two sulfated bi-antennary glycans were observed, which was not reported in the literature; these sulfated glycans were well resolved from their non-sulfated counterpart (Figure 4). The possibility of phosphorylation was excluded by alkaline phosphatase reaction (data not shown). In addition, MS/MS fragmentation was used to further confirm their identification (see Figure 5).
The glycan profile from both rFIX and pd-FIX is very complex. HPLC-based methods lack the resolution needed to identify and quantify various glycan forms. The ACQUITY UPLC System coupled with the ACQUITY UPLC BEH Glycans Column alone is able to achieve baseline separation of glycans that are different in mass and degree of sialylation; terminal sialic acid isomers are also well separated.
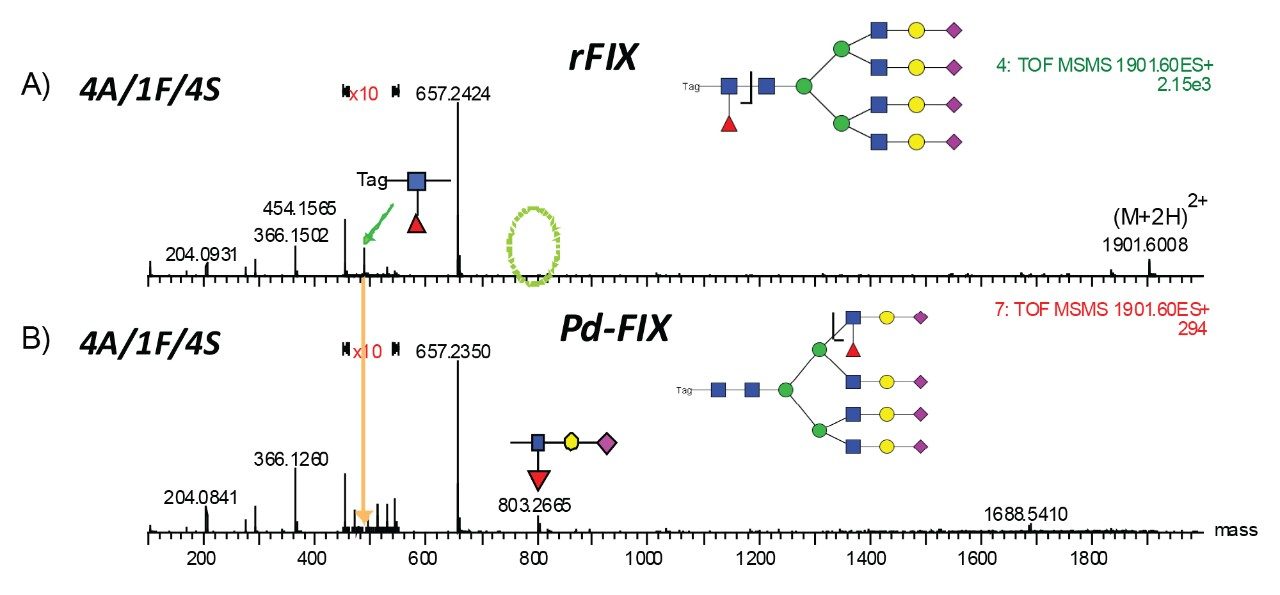
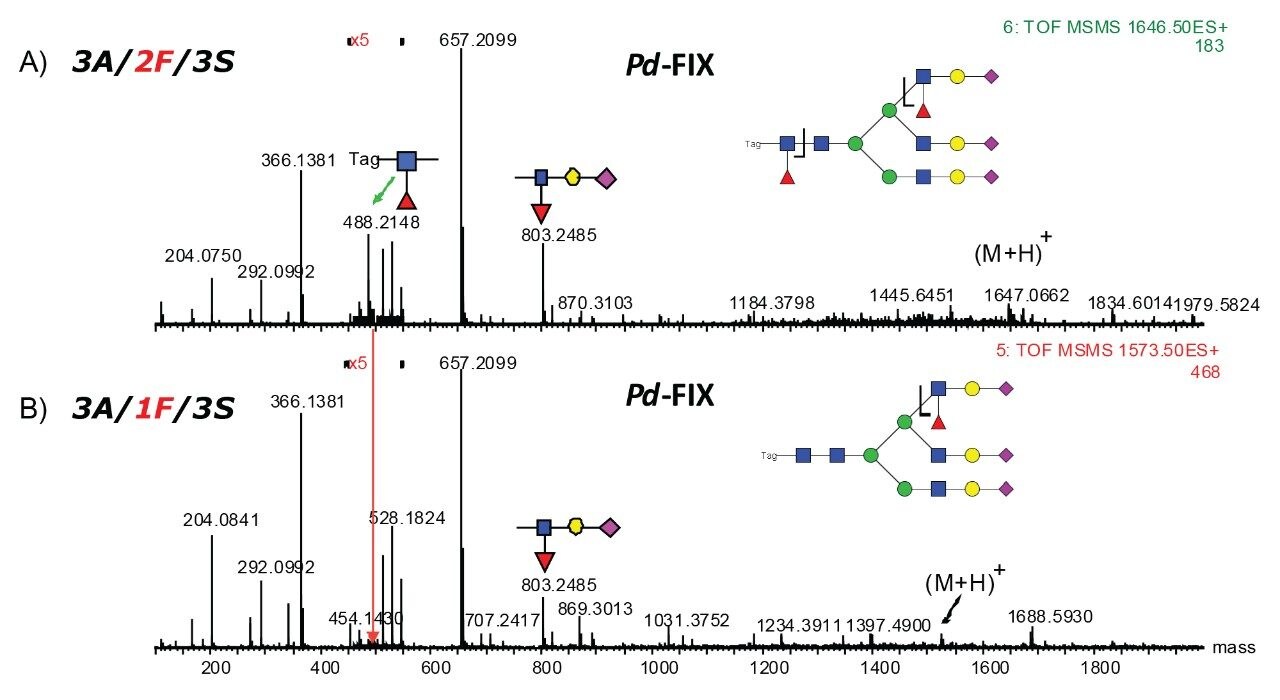
The accurate mass measurement from the Xevo QTof MS offers confident assignment of the glycans. MS/MS fragmentation along with SimGlycan Software gives further confirmation on glycan structure. Figure 6 shows that glycan 4A/1F/4S was observed from both rFIX and pd-FIX; the LC retention time is the same for the isobaric tetra-antennary glycans. However, the CID fragmentation showed distinct difference, which resulted from the fucose location on the glycans (Figure 5B). Figure 7 shows another example on how CID was used to differentiate isobaric doubly fucosylated glycans from both rFIX and pd-FIX glycans, again distinct fragment ions were used to differentiate the location of fucose. Biological influence caused by the location of fucose was documented in the literature.
Blood protein glycan characterization is known to be very challenging for scientists working in the biopharmaceutical field, since the glycans that attach to the protein backbone are highly heterogeneous and complex. Our solution for complex glycan separation and characterization is the UPLC/FLR/QTof MS analytical platform.
In addition to shortened run time enabled by ACQUITY UPLC technology, HILIC UPLC also offers significant improvement in peak resolution compares to conventional HPLC method; for example, positional sialic acid isomer separation is achieved, also the separation of sulfated and sialyated glycans were observed.
MS/MS fragmentation and database search using SimGlycan Software helped the glycan structure elucidation, adding more confidence to glycan structure assignment.
The ACQUITY UPLC/FLR/Xevo QTof MS analytical platform along with SimGlycan Software is a powerful and versatile tool for complex glycan characterization. Glycan profiling, mass confirmation, and structure elucidation can all be done in a single LC/MS system. High quality data are generated with shorter analysis and data interpretation time. This is a valuable tool for researchers working with glycoprotein drugs.
720004019, June 2011