In this study, we describe an optimized DDA method for peptide PTM analysis using the UNIFI Scientific Information System with the Vion IMS QTof MS especially for low abundant peptides with interference from co-eluting peptides. The high sensitivity and high dynamic range specifications of the Vion IMS QTof enable high quality MS/MS data for low abundant peptides and accurate quantitation. MSE combined DDA methods meet all the requirements in peptide mapping qualitatively and quantitatively.
Utilizing peptide mapping DDA workflow in UNIFI Scientific Information System with the Vion IMS QTof MS to improve MS/MS data quality for low abundant peptides from mAbs.
Data independent acquisition (DIA or MSE) and data dependent acquisition (DDA) are two widely used methodologies in biopharmaceutical laboratories for protein characterization. Waters QTof mass spectrometers and informatics solutions support both acquisition modes. In MSE method, mass spectrometers alternate scans equally between low and high collision energy (CE) to produce both precursor (low CE) and product ions (high CE) information. MSE benefits from Waters proprietary peak list generator (PLG) algorithm by aligning the retention time of a precursor and its fragment ions to generate both precursors and their corresponding product ion data,1 thus producing fragmentation data for all precursors under exact mass conditions without prior knowledge of the precursor ions. Because of the unique features in data acquisition and data processing, MSE is straightforward in method setup and provides both qualitative and quantitative information. In practice, MSE is typically used for routine peptide mapping experiments to accomplish the tasks of sequence confirmation and modification identification, and modification percentage calculation of PTMs (post-translational modifications). On the other hand, DDA, with the proper experimental set up, can be used as a complementary tool to obtain improved fragmentation data for low abundance peptides with PTMs to localize the modification site(s). This is especially beneficial for co-eluting peptides, which normally yield chimeric elevated energy mass spectra without precursor isolation. The main challenge is that the DDA method setup is not as straightforward as MSE. In this study, we investigate glycation of the NIST reference mAb as an example to demonstrate DDA method optimization for low level PTMs and highlight the advantage of DDA in obtaining more site-specific fragmentation and/or PTM localization.
NIST monoclonal antibody reference material (mAb) 8671 was diluted to 1 mg/mL in a denaturation buffer (7 M guanidine chloride, 0.2 M Tris, pH 7.5). Then, the diluted sample was reduced with 0.5 M DTT at 37 °C for 30 min and alkylated with 0.5 M iodoacetamide at room temperature for 20 min. Buffer exchange (to 0.1 M Tris, pH 7.5) of NIST mAb sample was performed using a NAP-5 column (GE Healthcare) prior to a fast tryptic digestion (30 min at 37 °C). The digested samples were stocked at -80 °C before LC-MS analysis. A total of 3 µg of tryptic digests were loaded onto ACQUITY UPLC BEH C18 Column for each injection for detecting the low abundance glycated peptides.
|
LC system: |
ACQUITY UPLC I-Class |
|
Analytical column: |
ACQUITY UPLC BEH C18 2.1 × 150 mm, 1.7 μm (P/N: 186003556) |
|
Column temp.: |
65 °C |
|
Mobile phase A: |
H2O with 0.1% formic acid |
|
Mobile phase B: |
Acetonitrile with 0.1% formic acid |
|
LC gradient: |
1–40% B in 150 min |
|
Acquisition mode: |
DDA |
|
MS system: |
Vion IMS QTof (Driver 2.0) |
|
Capillary voltage: |
2.5 kV |
|
Sampling cone: |
40 V |
|
Source offset: |
80 V |
|
Source temp.: |
100 °C |
|
Desolvation temp.: |
250 °C |
|
Cone gas flow: |
50 L/h |
|
Desolvation gas flow: |
600 L/h |
|
Reference mass: |
Leucine enkephaline [M+H]+ m/z 556.27658 |
|
MS to MS/MS Acquisition range: |
100–2000 m/z |
|
MS scan time: |
0.4 s |
|
MS/MS scan time: |
0.3 s |
|
Collision energy: |
6 eV |
UNIFI v1.8 SR 2 peptide mapping workflow
The detailed DDA method setup in the UNIFI Scientific Information System is demonstrated in Figure 1. A DDA experiment consists of many user-defined MS and MS/MS parameters in tabulation, as illustrated in the following screen captures. The details and the significance of key parameters and their effect on PTM identifications will be discussed in the following section. These settings include, but are not limited to, scan time, minimum acquisition intensity, maximum simultaneous MSMS acquisition, stop MSMS acquisition, and the dynamic peak exclusion.
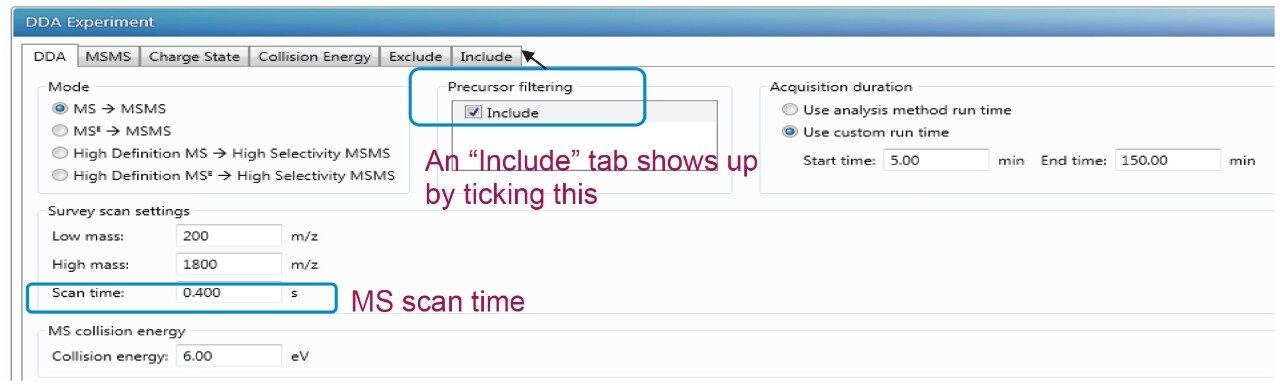
Experimentally measured duty cycles in DDA include one MS acquisition of mass/charge ratio (m/z) 100–2000, and the top most-intense precursor ions subjected to MS/MS analysis. The chromatographic gradient or the average peak width and the scan time decide the duty cycles in the DDA analysis. The longer scan time cumulates more ion intensity and provides higher data quality, but suffers a lower duty cycle which means fewer ions are selected and undergo MS/MS events.
This setting refers to the minimum ion intensity (Figure 2) required for a precursor ion in the full MS scan to be automatically selected for fragmentation in DDA mode. Increasing the signal threshold will result in lower number of acquired MS/MS spectra but the overall quality of each MS/MS spectrum can be improved. Decreasing the level of the signal threshold will result in a large number of MS/MS events, but the quality of the MS/MS spectra arising from low intensity ions may be insufficient for peptide identification. A threshold of 5000 to trigger MS/MS acquisition is found to be the optimal setting for low abundance PTMs experiment when the Vion QTof mass spectrometer is used. The threshold setting can be adjusted to match with the peptides with low ion intensities.
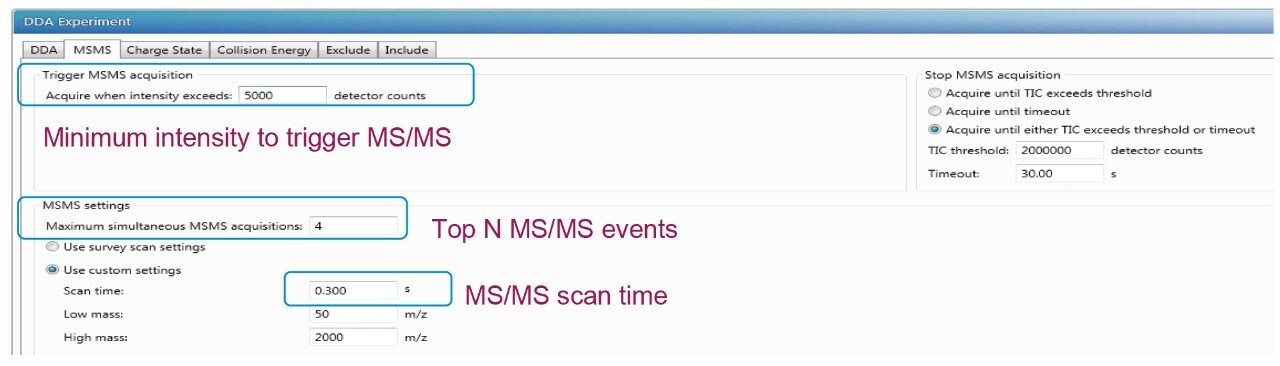
The setting means the “N” most abundant ions are selected for MS/MS analysis, shown in Figure 2. Increasing the number of MS/MS events would allow for the selection of lower-intensity ions but at the expense of increased cycle time. The increase in cycle time is not productive for fast-gradient conditions that produce narrow peak widths. For example, the cycle time increases to over 3.4 s with the selection of the top ten most-abundant ions with a fixed MS scan of 0.4 s and MS/MS scan of 0.3 s, this would not allow for multiple cycles to occur over the average 10-s-wide chromatographic peaks.
The MS/MS events are controlled by either the TIC counts or the timeout depending on which parameter is reached first. Timeout is the length of time the m/z is eligible for MS/MS acquisition before it is no longer subjected to further fragmentation. A value of 30 s for MS/MS acquisition, shown in Figure 2, was selected to correspond with observed average peak width at base for PTMs identification. The TIC counts are critical in identification of low intensity precursor ions. To allow the low intensity precursor ions to be subjected to MS/MS fragmentation, it is important to have lower TIC counts to preventing the high abundance ions staying too long in MS/MS mode. In this case, a TIC of 2e6 was selected for PTMs analysis.
With dynamic peak exclusion enabled, an m/z ion is placed on an exclusion list and will be excluded from repeated MS/MS analysis for a specified time, called exclusion duration. When dynamic exclusion is disabled, repeated MS/MS scans of the more intense precursor ions will be generated. In identifying low abundance PTMs, the dynamic exclusion is normally enabled to minimize repeated sequencing of peptides and allow for lower abundance ions to undergo fragmentation. Reduction of dynamic exclusion time improved sequence coverage and increased the number of matched MS/MS spectra; however, this also led to an increased number of redundant MS/MS spectra collected. The preferred dynamic exclusion time for the current study was 150 s (Figure 3).
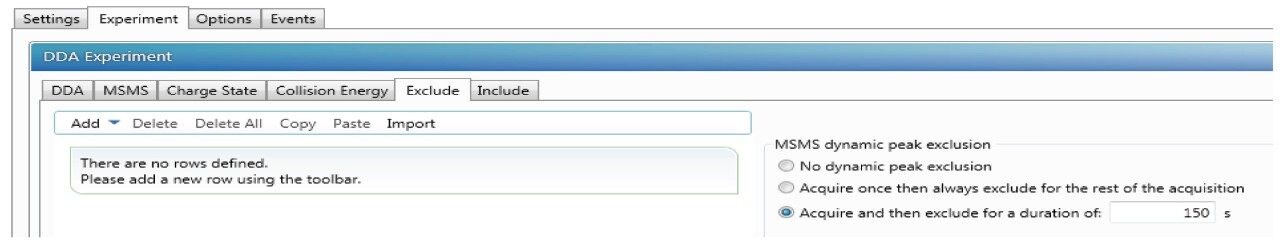
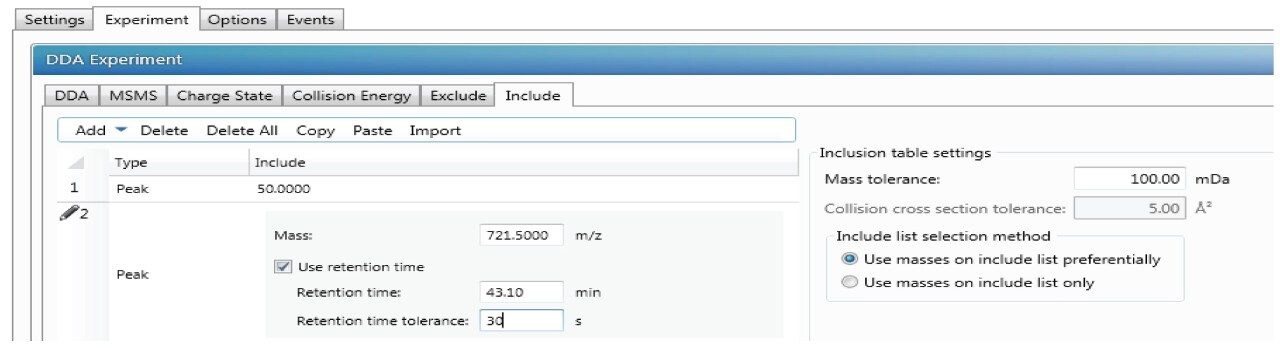
DDA acquisitions are typically performed in an untargeted mode and can be complemented with include and/or exclude lists as well. Inclusion list is enabled by selecting “Include” under DDA tab as shown in Figure 1. In the “Include” tab, certain m/z and RT or certain mass range could be added with tolerance time window as shown in Figure 4.
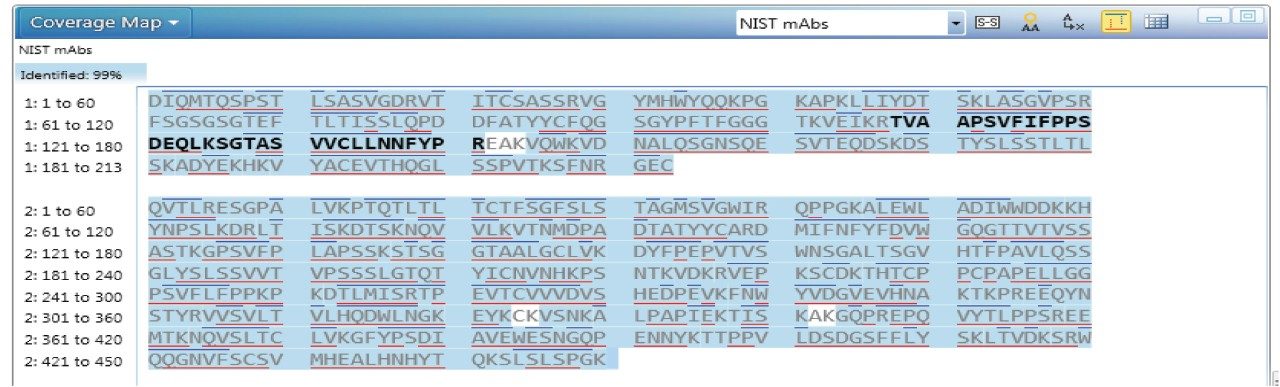
In this study, a fast 30 min proteolytic digestion was performed after the reduction and alkylation of NIST mAbs to minimize sample preparation related artifacts. Triplicate analysis of the NIST mAb tryptic digests was undertaken via the peptide mapping workflow in UNIFI. Sequence coverage of 99% was observed, as shown in Figure 5, based on the following strict criteria set: 1) identified peptides must have at least 3 confirmatory fragment ions (b or y ions); 2) up to 2 missed cleavages are allowed but no semi-tryptic peptides considered; 3) mass accuracy is within 10 ppm for precursors; 4) no neutral loss of NH3 or H2O, and no in-source fragments are counted.
With naturally low abundance in NIST mAbs, glycated peptides usually show very little fragmentation across the peptide bonds in an MSE experiment. In this application note, we demonstrate an optimized DDA method to identify the glycated peptides using NIST mAb RM 8671 due to its high glycation level.
All unmodified Lys residues are susceptible to glycation in theory. However, the rate of individual modification varies depending on local sequence and higher order structure.2 The presence of glycation prevents trypsin and lys-C cleavage at the modified residue.3 Thus, up to 2 missed cleavages were considered during peak searching in UNIFI. In the DDA method, a lower intensity threshold of 5000 was used to trigger an MS/MS event. The low trigging threshold allows more low abundance precursor ions to be subjected to MS/MS fragmentation. In addition, a TIC threshold of 2e6 and a timeout of 30 sec were enabled to prevent the same high intensity precursor ions from undergoing repeated fragmentation. With these DDA parameters, more fragment ions were observed for low abundance ions such as the glycated peptides compared to MSE. The identified glycated peptides with modification levels from 0.05 to 4.77% cover 23 lysines in NIST mAbs. The identified glycated sites and the site occupancy are calculated by the MS response of glycated peptide divided by the sum of glycated peptide and unmodified peptide. An equation is shown below using a representative peptide. The results are listed and compared with the published result4 at Table 1.

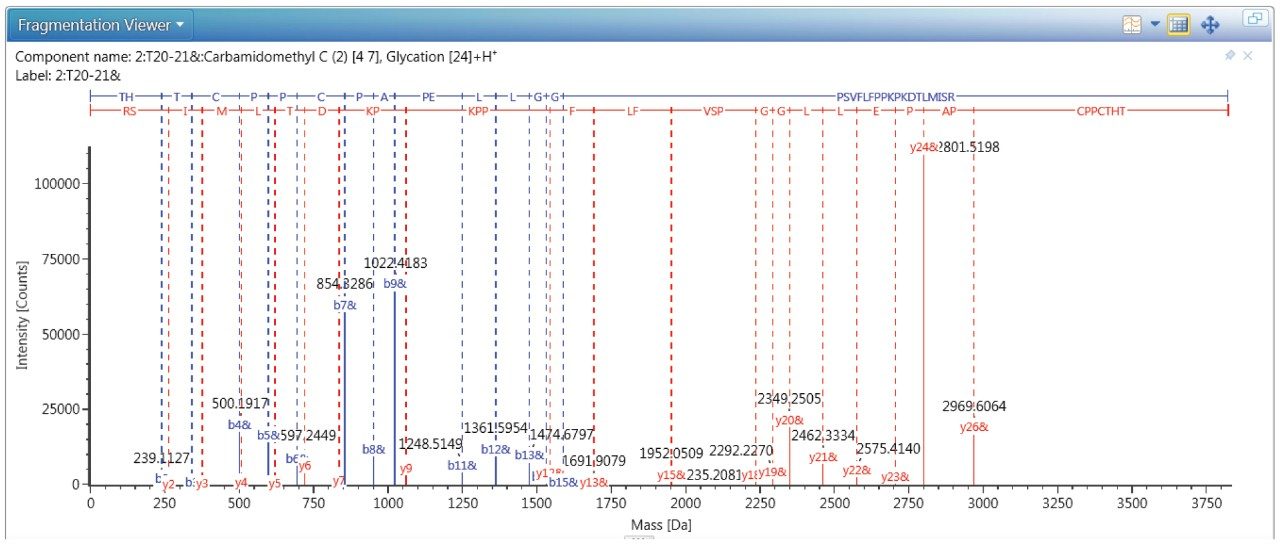
The MSMS spectrum of one representative peptide (THTCPPCPAPELLGGPSVFLFPPK249PK251DTLIMISR) with two lysine resides of K249 and K251 is shown in Figure 6, the MS/MS confirmed the glycation is site-specific to residue 249 and the site occupancy is 0.32%.
In this study, we describe an optimized DDA method for peptide PTM analysis using the UNIFI Scientific Information System with the Vion IMS QTof MS especially for low abundant peptides with interference from co-eluting peptides. The high sensitivity and high dynamic range specifications of the Vion IMS QTof enable high quality MS/MS data for low abundant peptides and accurate quantitation. MSE combined DDA methods meet all the requirements in peptide mapping qualitatively and quantitatively.
720006134, November 2017