This is an Application Brief and does not contain a detailed Experimental section.
This technology brief demonstrates the newly integrated data-processing capability of Waters High Definition Imaging Software (HDI), version 1.4. Datasets of different mass spectrometry modalities (such as DESI and MALDI) are processed consecutively with user-defined settings.
The workflow for processing MS imaging data has been simplified and is entirely performed within HDI, avoiding the need for further use of MassLynx Software. Simultaneous batch processing of multiple datasets with different experiment types is possible. Users can process the same imaging dataset under a variety of different parameters, with the option to create a new file each time. Finally, HDI 1.4 improves mass accuracy with a new internal lock mass function implemented for MALDI and DESI datasets.
Mass spectrometry imaging (MSI) is gaining popularity in the field of analytical science. Waters offers both MALDI- and DESI-based imaging configurations on the SYNAPT G2-Si Mass Spectrometer, as well as DESI-based imaging configurations on Xevo G2-XS instruments. As MSI becomes an established and widespread technique, software that is used for acquisition and subsequent manipulation of datasets must meet the requirements of end users.
Waters has developed an improved software solution: High Definition Imaging (HDI) Software, version 1.4. This software incorporates a new user interface to mine the processed MSI data, and is specifically designed to be user-friendly and intuitive. Many features and capabilities have been added in order to enable rapid, convenient data management.
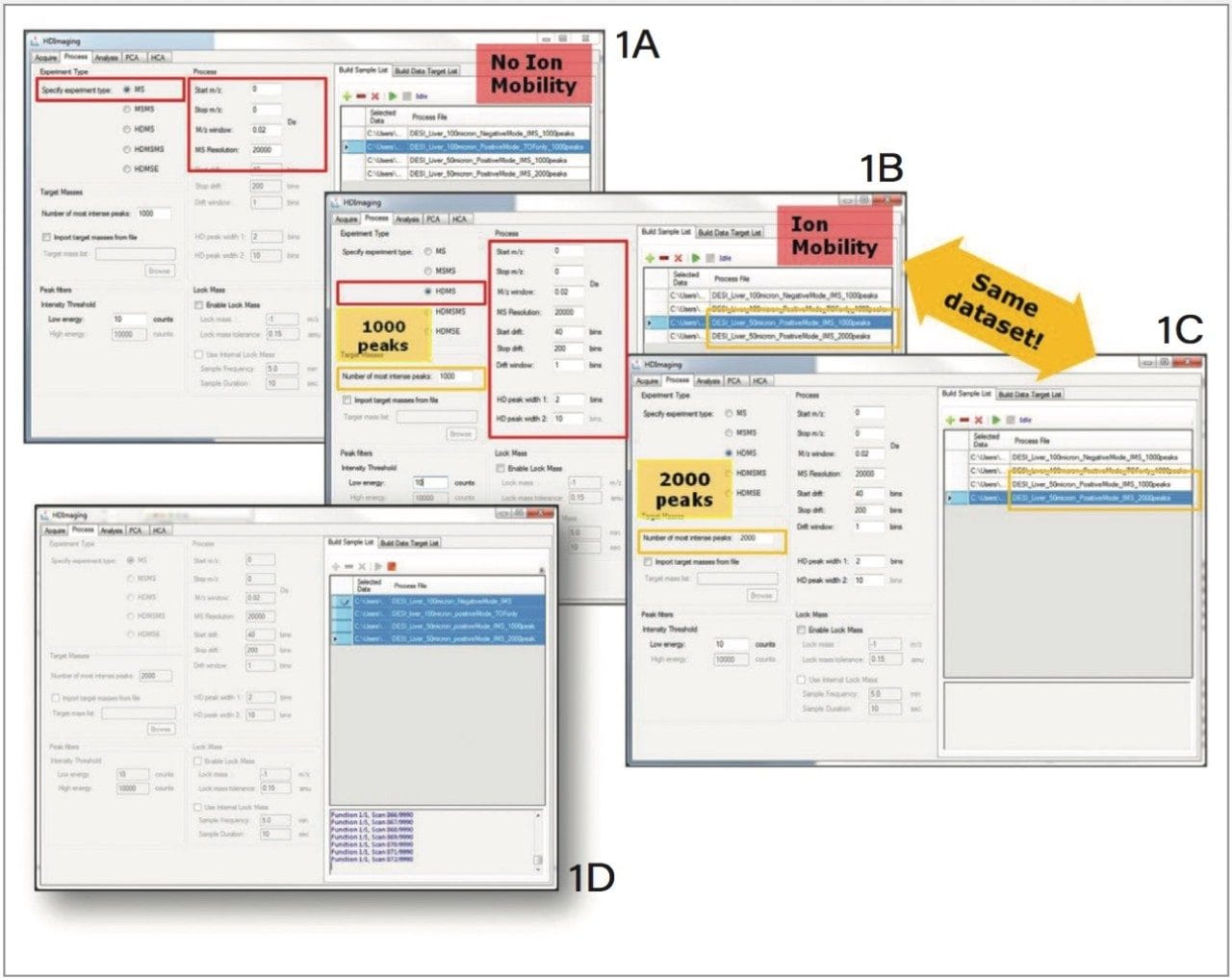
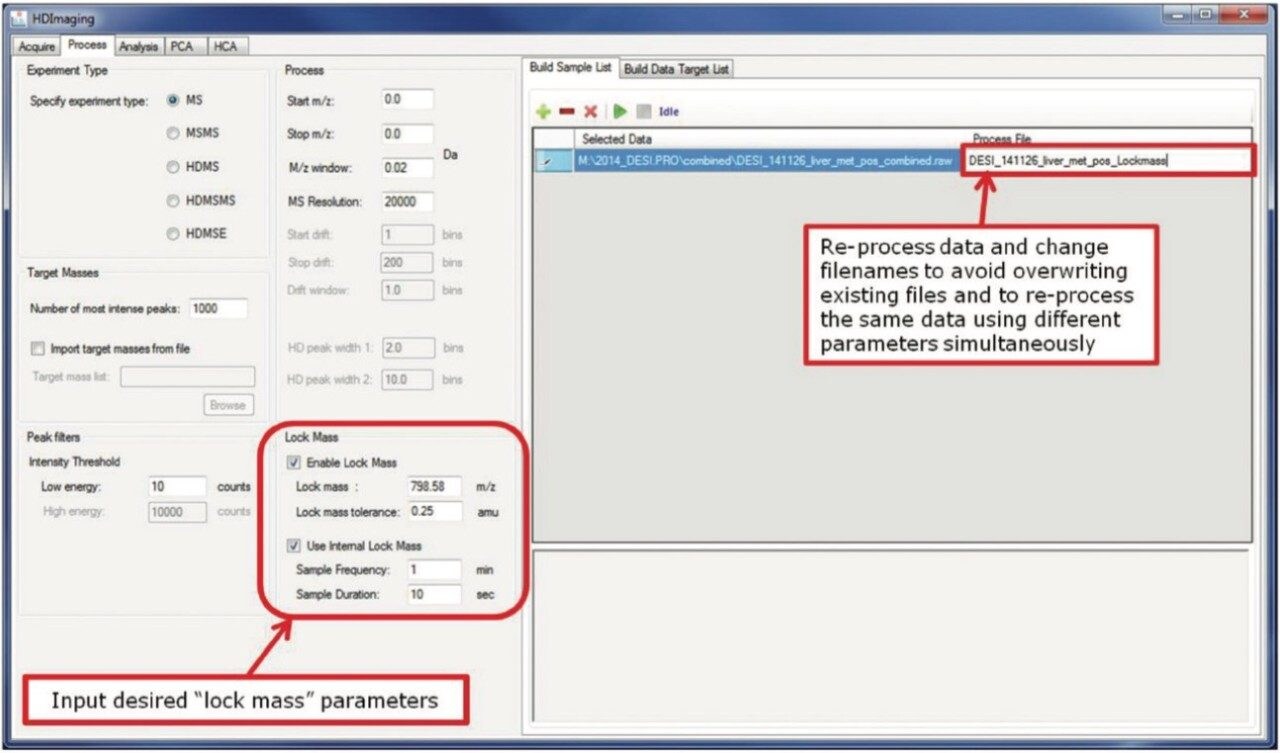
With HDI 1.4 it is possible to process datasets of different experimental types. The individual datasets are initially loaded into the “Build Sample List” tab on the right, and their specific processing parameters are set up in the left-hand panel. This is done sequentially for each individual dataset loaded into the sample list. In addition to simultaneous processing of different datasets, the same dataset may be processed multiple times with alternative processing parameters. To support this function, it is also possible to change the filename of the newly created processing file — thereby avoiding an overwrite of existing files (Figure 2).
Raw data is now processed directly in HDI 1.4 (Figure 1D), so there is no need to use the MassLynx Application Manager, as was previously the case with this software. Also shown in Figure 1D, after processing is initiated, progress can be viewed in the live pane at the bottom of the screen. While data processing is running, it is possible to continue the mining and review of other (previously processed) datasets in the “Analysis” tab.
In addition to the external lock mass traditionally implemented for Waters MALDI imaging experiments, a new internal lock mass function has been implemented; this allows for reliable lock mass correction in both MALDI and DESI experiments.
Regardless of the method, the lock mass m/z is user-defined, along with the tolerance within which the lock mass peak is expected to appear. When using an internal lock mass, the user can input the desired sampling frequency and duration. In the case of the example shown in Figure 1, the software will look for the specified lock mass at one minute intervals for a duration of 10 seconds throughout the dataset.
HDI Software, version 1.4, offers a number of improved features to benefit the researcher.
The workflow for processing MS imaging data has been simplified and is entirely performed within HDI, avoiding the need for further use of MassLynx Software. Simultaneous batch processing of multiple datasets with different experiment types is possible. Users can process the same imaging dataset under a variety of different parameters, with the option to create a new file each time. Finally, HDI 1.4 improves mass accuracy with a new internal lock mass function implemented for MALDI and DESI datasets.
720005773, July 2016