This application note shows high resolution UPLC-MS for the separation and identification of phospholipids.
Phospholipids, a class of lipids that contain phosphorus and are categorized by specific polar head groups, have received much attention in many scientific and medical fields. A metabolic disorder of the fatty acid components of phospholipids has been shown to correlate with the incidence of diabetes mellitus. This disease effects 10 percent of the world’s population over the age of 60, and consumes a large proportion of health care expenditures.1
In addition to the importance phospholipids have in the study of certain disease states, this class of lipid has many commercial uses. Cosmetics, detergents and drug delivery vehicles all incorporate phospholipids into their formulations.
However, limitations exist with the current methods used in the analysis of phospholipids, primarily due to the structural diversity of these molecules. UltraPerformance LC (UPLC), a technique that greatly improves resolution over traditional HPLC analysis, has shown great potential in the separation of complex samples.2 In this application note, we show high resolution UPLC-MS for the separation and identification of phospholipids.
Standards were obtained from Avanti Polar Lipids, Inc. (AL, USA). Rat plasma was obtained from Equitech-Bio Inc. (TX, USA) and precipitated with acetonitrile, 2:1.
|
LC system: |
Waters ACQUITY UPLC System |
|
Mobile phase A: |
20 mM ammonium acetate, pH 5.0 |
|
Mobile phase B: |
Acetonitrile/acetone (9:1) |
|
Gradient: |
35 to 95% B/10 min |
|
Flow rate: |
600 μL/min |
|
Column temp: |
60.0 °C |
|
Column: |
ACQUITY UPLC BEH C8, 2.1 x 100 mm, 1.7 μm |
|
MS system: |
Waters LCT Premier XE Mass Spectrometer |
|
Ionization mode: |
ESI Positive |
|
Capillary voltage: |
3,200 V |
|
Cone voltage: |
35 V |
|
Desolvation temp: |
400 °C |
|
Desolvation gas: |
800 L/Hour |
|
Source temp: |
120 °C |
|
Acquisition range: |
100 to 1,000 m/z |
|
Acquisition rate: |
0.095 sec |
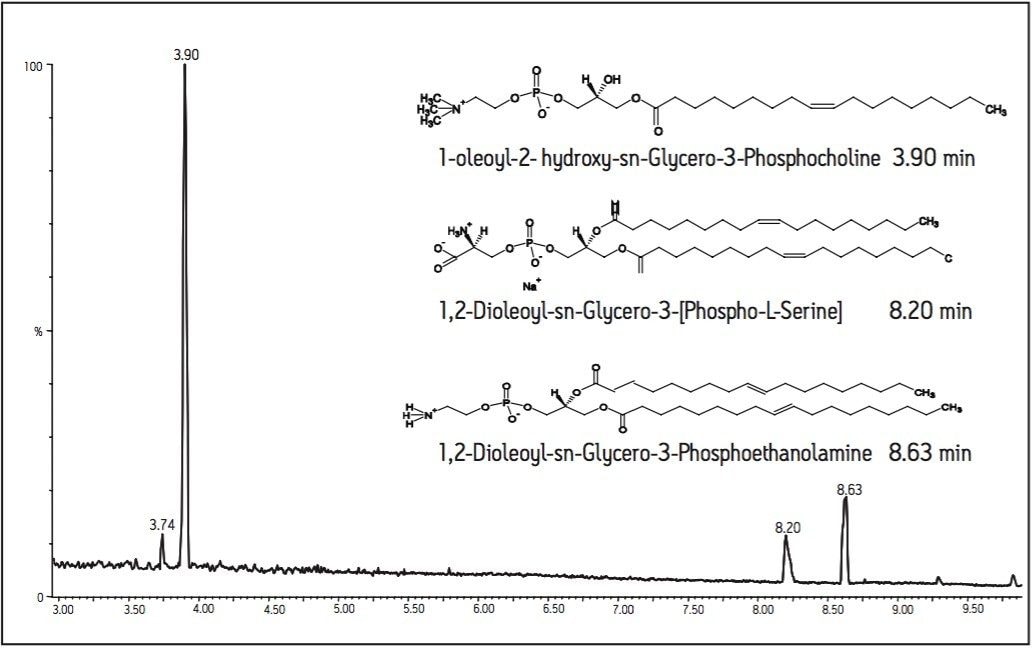
The UPLC-MS separation of three phospholipids standards can be seen with their corresponding chemical structures in Figure 1. The UPLC-MS system generated a high resolution chromatogram and good peak shape with little (if any) peak tailing. Average peak widths at the base of 3 seconds yielded a peak capacity of 200 in 10 minutes. This same method was then applied to protein precipitated rat plasma samples.
Here again, we can see that UPLC-MS produced a high peak capacity separation (Figure 2). Lysophospholipids (LPL) and phospholipids (PL) are known to exist in the supernatant of protein precipitated plasma.3 Most of the peaks seen in this figure can be separated into two distinct groups. The first group includes those that elute between 3 and 5 minutes, representing a majority of the LPL present in the sample.
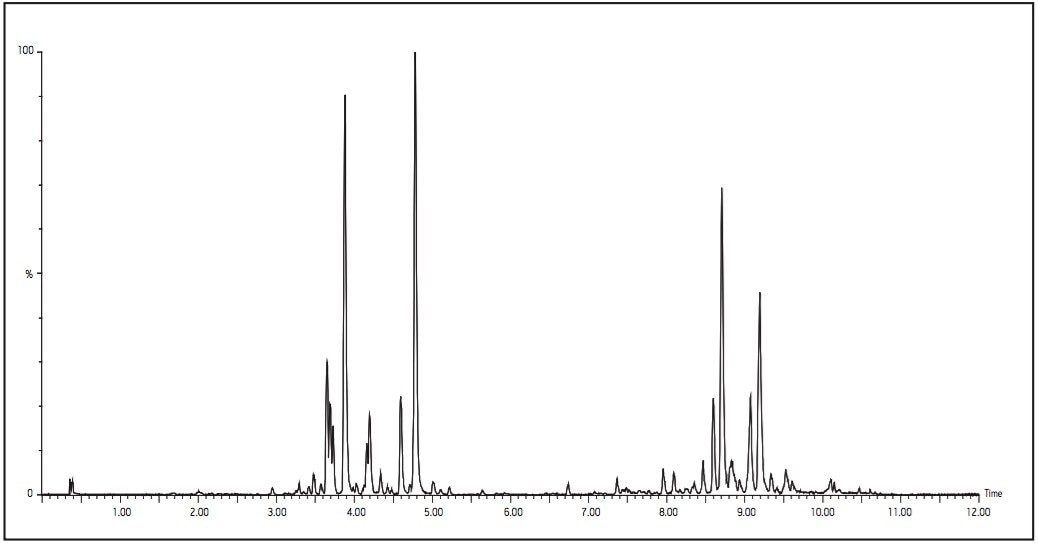
The second group includes those that elute between 7 and 10 minutes, representing a majority of the PL present in the sample. Further experiments were conducted to determine these peaks as LPL and PL of various fatty acid compositions. These experiments further identified the majority of these LPL and PL to contain the choline head group (data not shown).
In this application note, we demonstrate the ability of UPLC coupled with the LCT Premier XE to separate different species of phospholipids. Good peak shape with average widths of three seconds at base yielded high peak capacity separations. The methodology was further applied to the analysis of complex phospholipid species extracted from mammalian plasma, again producing a high peak capacity separation.
These results show that the high resolution capability of UPLC-MS to be an exceptional platform for targeted lipid analysis of phospholipids, as well as for complex lipidomic studies.
720001872, February 2007