
In this application note, we optimized the trypsin digestion step for the development of a sensitive LC-MRM assay for trastuzumab in human serum.
The analytical workflow of trypsin digestion can be optimized to develop a successful multiple reaction monitoring (MRM) assay. Results include higher assay sensitivity, reduced sample preparation time, and higher assay reproducibility.
Mass spectrometry-based quantification of therapeutic proteins uses a surrogate peptide as a stoechiometric representative of the protein which is cleaved. One of the challenges encountered in the quantification of therapeutic proteins in biological fluids (for example serum, plasma, and urine) is to find a suitable internal standard for each protein analyte. Isotopically labeled protein standards have been used successfully for quantification of mAbs,1-3 but these standards are expensive and time-consuming to produce.
Since protein quantification is performed at the peptide level, another alternative is to use relatively inexpensive isotopically labeled peptides as peptide IS.4-7 To account for trypsin digestion efficiency, 13C15N-isotopically labeled cleavable peptides (extended peptides) were introduced in 2004.5
Regardless of the nature of the IS, both quantification approaches rely on the digestion of the protein sample with a specific enzyme (such as trypsin, Lys C). Protein digestion has been recognized as the major source of variability in the analytical workflow4-9 which has to be carefully optimized. In addition, when the whole digest approach is implemented, using peptide IS for quantification, there is an additional concern related to the ability of the digestion enzyme to cleave with the same efficiency as the therapeutic protein as well as the isotopically labeled peptide IS, in the presence of the biological matrix.
In this application, trypsin digestion optimization is required for developing a successful MRM assay. Generally, some of the benefits of trypsin digestion optimization include higher assay sensitivity, shorter sample preparation times, and higher assay reproducibility.
Here, we used a therapeutic mAb to investigate the trypsin digestion efficiency relative to the whole digest quantification method. Two digestion parameters, protein to trypsin ratio and digestion time, were optimized. Additionally, the reproducibility of the entire sample preparation protocol was assessed.
Trastuzumab (herceptin) is a humanized IgG1 kappa monoclonal antibody (mAb). The antibody was produced through genetic engineering10,11 by joining the constant regions of the human monoclonal antibody with the complementaritydetermining regions (CDRs) of a mouse monoclonal antibody able to bind human epidermal growth factor receptor 2 proteins (HER2) receptors. These HER2 receptors belong to a family of human oncoproteins expressed in approximately 25% of invasive breast cancers. Trastuzumab was approved in 1998 by the U.S. Food and Drug Administration (FDA) for the treatment of HER2-overexpressing breast cancers.
In this application note, we optimized the trypsin digestion step for the development of a sensitive LC-MRM assay for trastuzumab in human serum.
|
System: |
ACQUITY UPLC I-Class |
|
Column: |
BEH300 C18 2.1 x 150 mm, 1.7 μm |
|
Column temp.: |
35 °C |
|
Flow rate: |
0.3 mL/min |
|
Mobile phase A: |
0.1% (v/v) formic acid (FA) in water |
|
Mobile phase B: |
0.1% (v/v) FA in acetonitrile |
|
Linear gradient: |
0% to 35% B in 10 min |
|
Run time: |
15 min |
|
Mass Spectrometer: |
Xevo TQ-S |
|
Mode: |
MRM positive ion electrospray |
|
ESI potential: |
3.5 kV |
|
Cone voltage: |
28 V |
|
Source temp.: |
120 °C |
|
MS1/MS2 isolation window: |
0.75 Da (FWHM) |
|
Collision energy: |
28 eV |
Four MRM transitions were monitored continuously throughout the LC-MRM assay using a dwell time of 50 ms: two MRMs monitored the endogenous signature peptide FTISADTSK from trastuzumab (485.2 → 721.4 for peptide quantification; 485.2 → 608.3 for peptide confirmation), while the other two MRM channels monitored the corresponding 13C15N-isotopically labeled internal standard peptide FTISADTSK (489.2 → 729.4 for peptide quantification; 489.2 → 616.3 for peptide confirmation).
Aliquots containing 40 μL of human serum were dispensed in Low-Bind Eppendorf vials and digested with trypsin, following the procedure described below, to produce 200 μL of human serum digest per vial. As shown in Figure 1, the digestion protocol involved sample denaturation (with 0.05% RapiGest at 80 °C for 10 min), disulfide bond reduction (in the presence of 20 mM dithiothreitol (DTT) for 60 min at 60 °C), and cysteine alkylation (with 10 mM iodoacetamide (IAM) for 30 min at room temperature in the dark). After adding the internal standard peptide (the 13C15N-isotopically labeled extended peptide GRFTISADTSK), samples were digested with porcine trypsin (Sigma catalog no T-6567) under different experimental conditions. In one experiment, the amount of trypsin was varied, so that the ratio between the substrate (total serum proteins) and the digestion enzyme could be altered. Five digestion ratios (10:1, 20:1, 30:1, 50:1, and 100:1) were tested. In another experiment, the substrate to enzyme ratio was kept constant (30:1), and the digestion time was varied. Aliquots were taken after 15 min, 30 min, 1 h, 3 h, 6 h, and 16 h (overnight). The digestion was stopped by adding TFA (2 μL), and samples were incubated for 30 min at 37 °C to decompose RapiGest. The supernatant was recovered from each sample, following centrifugation at 12,000 rpm (10 min), and 10 μL of sample was injected onto the LC-MS instrument.
Figure 1 shows a general analytical workflow with no analyte pre-fractionation, developed for the quantification of therapeutic proteins in serum, and tested for the quantification of trastuzumab in human serum. After spiking the mAb in serum, the protein mixture was enatured, reduced, and alkylated, as ndicated in steps 2 through 4 in Figure 1 (also see the Experimental section). The extended 13C15N sotopically abeled peptide GRFTISADTSK was spiked at a concentration of 100 nM, and digested with trypsin. The sample as diluted four old during digestion, making the final concentration of the peptide IS 25 nM. Trypsin is the referred digestion enzyme for protein ioanalysis applications for several reasons including: it is easily vailable, well characterized, and has very reproducible cleavage sites (K/R).

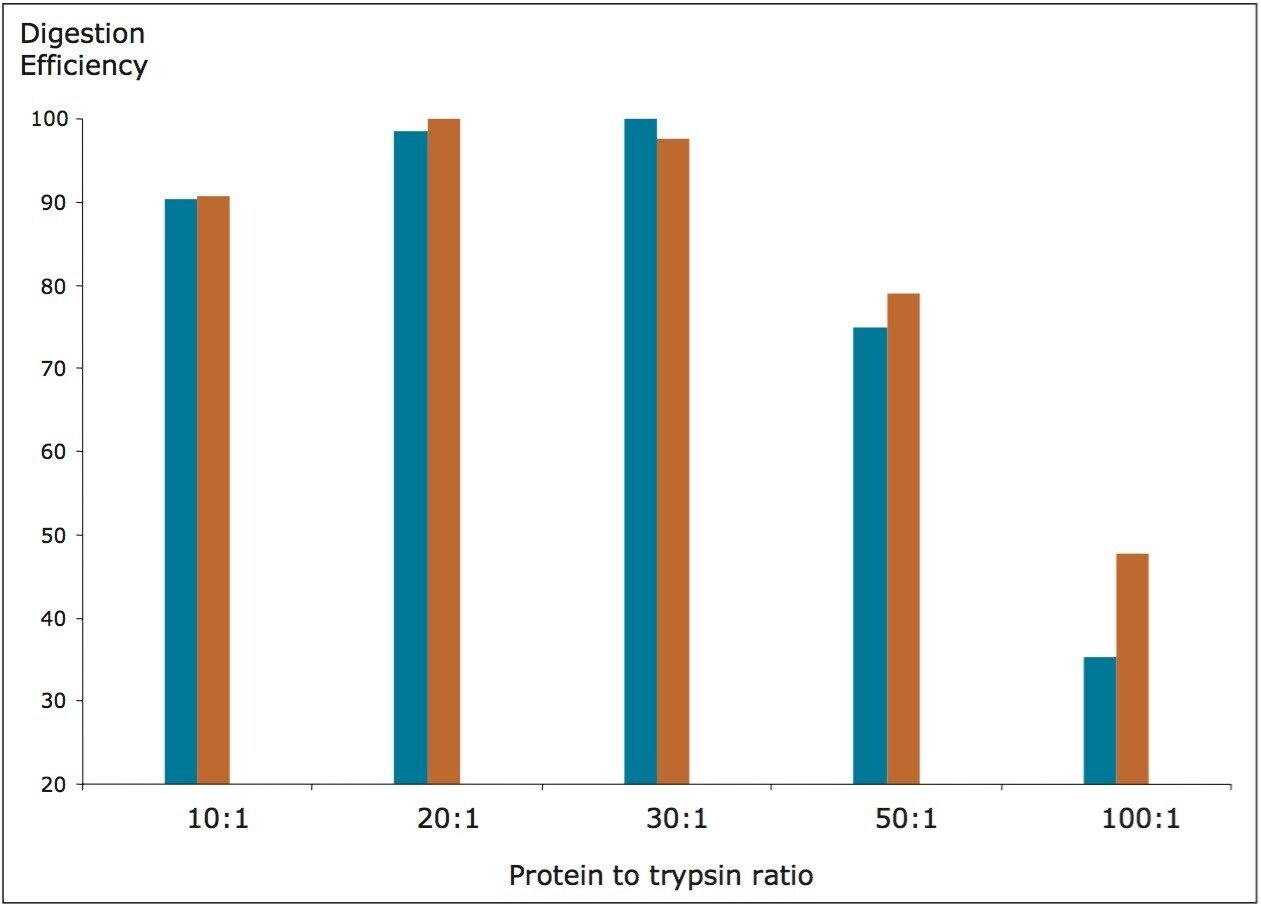
Two digestion parameters were investigated including: protein to trypsin ratio and digestion time. In one experiment, the amount of trypsin was varied so that the ratio between the substrate (total serum proteins) and the digestion enzyme could be altered. Five digestion ratios (10:1, 20:1, 30:1, 50:1, and 100:1) were investigated with the results shown in Figure 2. Digestion efficiency was calculated from the normalized averaged peak areas (n=4) obtained from the MRM chromatograms of the native FTISADTSK peptide (originating from spiked trastuzumab, MRM transition: 485.2 → 721.4), and the corresponding isotopically labeled analogue (FTISADTSK, 489.2 → 729.4).
According to the data shown in Figure 2, for substrate to enzyme ratio of up to 30:1, there are no significant differences between the digestion of the analyte (trastuzumab) and the digestion of the peptide substrate (extended peptide IS). However, at ratios above 30, trypsin becomes more efficient at digesting the smaller substrate, to the detriment of the therapeutic protein. Clearly, ratios lower than 30:1 should be used to avoid quantification errors due to the incomplete digestion of the analytical target. A decrease in trypsin efficiency at low substrate to trypsin ratios (for example 10:1) is typically associated with a higher amount of trypsin auto-cleavage at the expense of protein substrate cleavage.
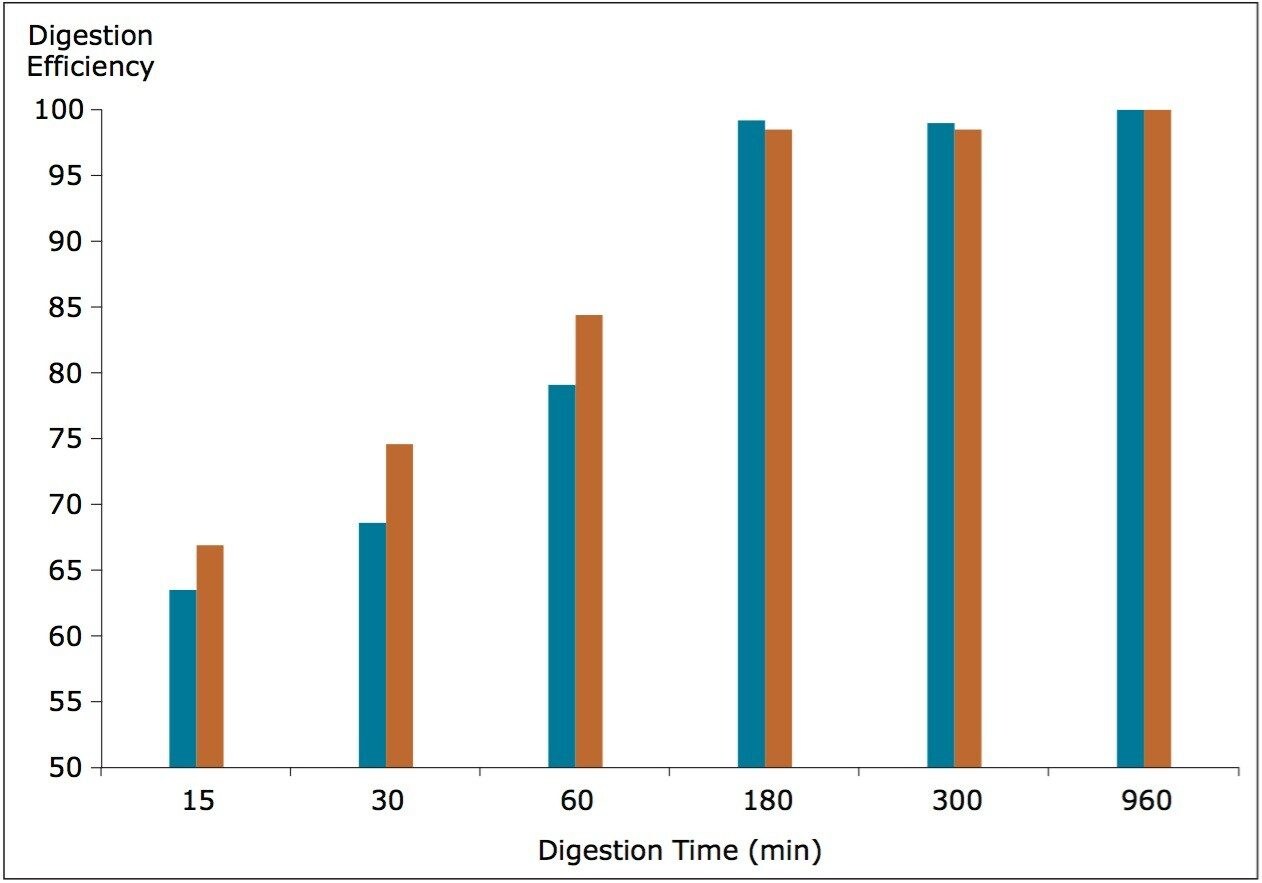
In addition to the substrate to trypsin ratio, the digestion time was evaluated. The substrate to enzyme ratio was kept constant (30:1) while the digestion time was varied. Aliquots were taken after certain digestion times [15 min, 30 min, 1 h, 3 h, 6 h, and 16 h (overnight)], and the digestion was stopped by adding TFA. Digestion efficiency was again calculated from the normalized averaged peak areas of FTISADTSK peptides, as shown in Figure 3. The optimum digestion time was 3 h with no signal increase observed after that point in time.
As shown in Figure 3, the reproducibility of the entire analytical workflow was tested at a lower concentration: 5 nM trastuzumab spiked in human serum with 5 nM 13C15N-isotopically labeled extended peptide added as an IS. Figure 4 shows the average peak areas obtained for five replicate digestions. While the peak area RSD was approximately 10% for individual MRM traces, the %RSD for the 12C/13C peptide ratio (used for quantification) was greater than 6%.
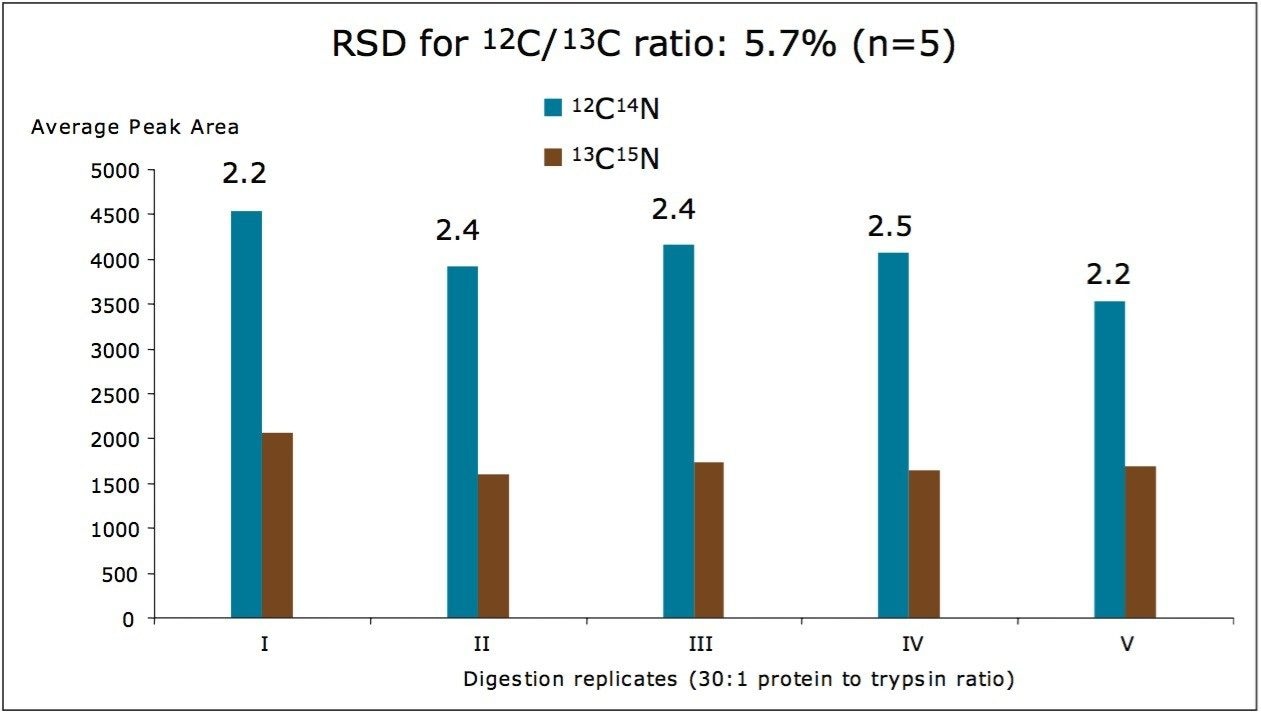
Even though the analyte trastuzumab and the peptide IS were spiked into human serum at the same concentration (5 nM), the ratio between the peak areas of the native peptide and the isotopically labeled peptide was approximately 2:1 because the therapeutic protein generates two peptide molecules for each molecule of digested mAb. Since the calculated 12C/13C peptide ratio is close to the expected value (2:1), it is clear that the trypsin digestion efficiency was minimally affected by the type of the substrate (150 kDa therapeutic mAb versus 1 kDa peptide IS). The whole digest approach can be employed for quantification of therapeutic proteins in serum.
A general workflow for the quantification of therapeutic proteins in serum without analyte pre-fractionation was tested to determine the quantification of trastuzumab in human serum.
720004507, November 2012