Advanced Oligonucleotide Characterization Using Multi-Reflecting Time-of-Flight Technology
Jonathan Fox1, Ying Qing Yu1, Scott J Berger1, Nick Pittman1
1 Waters corporation, United States
Published on May 02, 2025
Abstract
LC-MS has become a widely used analytical tool for oligonucleotide full length product (FLP) identity confirmation and impurity analysis due to its high sensitivity and selectivity. It provides the ability to perform comprehensive characterization for both the FLP and a wide range of impurities. This application brief demonstrates the use of the ACQUITY™ Premier UPLC System coupled to the Xevo™ MRT Mass Spectrometer for an antisense oligonucleotide analysis. The INTACT Mass and CONFIRM Sequence Applications within waters_connect™ software platform were employed for data processing, providing comprehensive insights into the GEM 91 oligonucleotide and its associated impurities from the intact mass profile and data independent fragmentation data. GEM 91 is a phosphorothioate antisense oligonucleotide utilized as a therapeutic agent in the treatment of AIDS. The results demonstrated consistent low to sub-ppm mass accuracy, high sensitivity, and high mass resolution that enabled mass confirmation of the FLP and confident detection of multiple product related synthetic impurities.
Benefits
- Low- to sub-ppm mass accuracy coupled with high levels of sensitivity and dynamic range facilitates in-depth oligonucleotide characterization and detection of subtle differences and modifications
- Efficient intact mass data analysis workflows combining automated peak detection, deconvolution, mass confirmation and purity analysis calculations, enables higher throughput analysis with reduced effort spent on data review, and less concerns over data transcription and manual calculation errors
- Automated confirmation of data-independent oligo fragmentation data provides greater efficiency of assessing oligonucleotide variation and avoids the human error and data integrity challenges of 3rd party spreadsheet and informatics tools
- Integrated compliance-ready app-based workflows for streamlined acquisition, processing, and review of oligonucleotide data enables more people in an organization to be successful with oligonucleotide characterization, and limits training required to be proficient with the analysis
Introduction
Oligonucleotide therapeutics are typically generated using synthetic chemistry-based methodologies, and can exhibit a range of synthesis related impurities, as well as degradation products that can affect the safety, stability, and efficacy profile of the molecule. Initial characterization and continued monitoring of product variation is essential for commercializing an effective product and demonstrating a resilient process to produce them. LC-MS has been readily adopted for both tasks, as it can confirm molecule identity, detect and assign sites of variation, and track those product variants across product and process development.
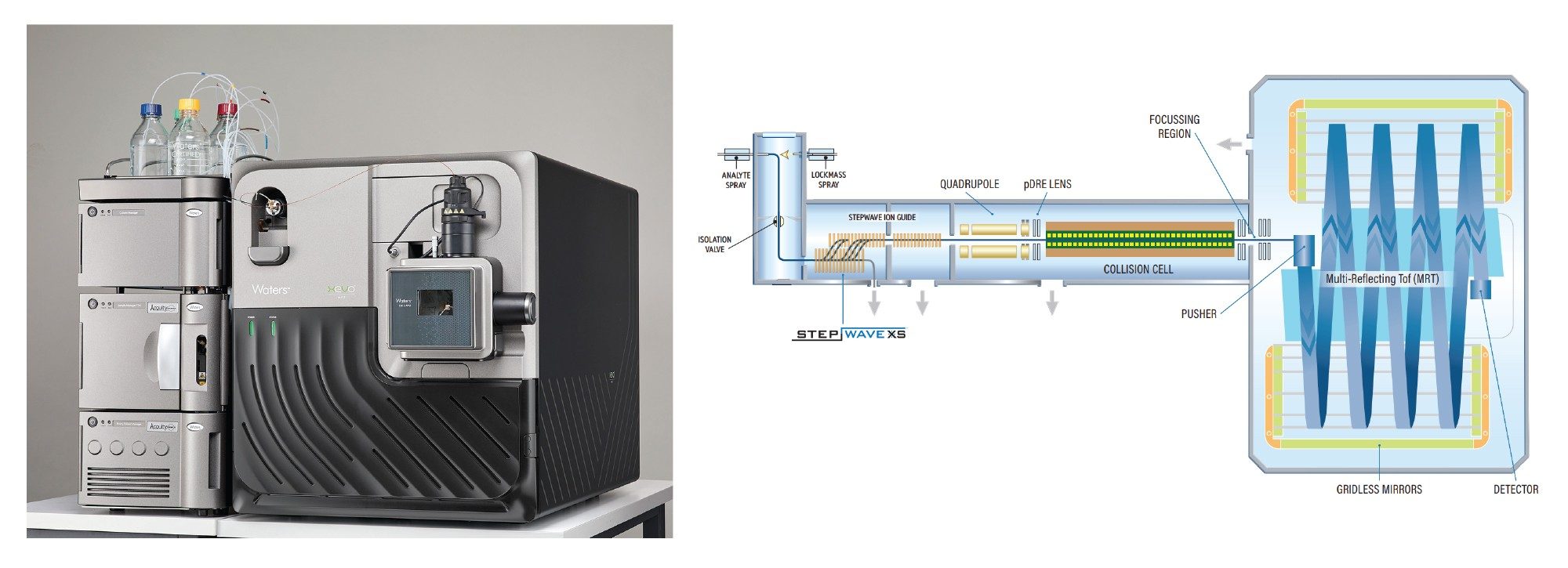
The Xevo MRT MS System (Figure 1) uses a novel multi-reflecting time-of-flight (TOF) geometry and instrument design to deliver exceptional sensitivity, dynamic range, mass resolution and mass accuracy for complex analytical tasks. The Xevo MRT MS achieves sub-ppm mass accuracy and a mass resolution of up to 100,000 FWHM, this combination of mass accuracy, sensitivity and mass resolution enables detailed oligonucleotide characterization. The Xevo MRT MS can detect and characterize complex and low intensity impurities which can significantly impact the efficacy and safety of oligonucleotide based biopharmaceuticals.
Following Ion-pairing reversed phase LC-MS data acquisition, data processing for intact oligo mass confirmation and impurity profiling can be conducted in the waters_connect INTACT Mass App. The INTACT Mass App employs sophisticated algorithms for LC peak detection and automated deconvolution of oligonucleotide spectra, providing rapid full-length product and impurity assignments as well as the required metrics (mass accuracy and abundance) to support quantitative impurity calculations.1
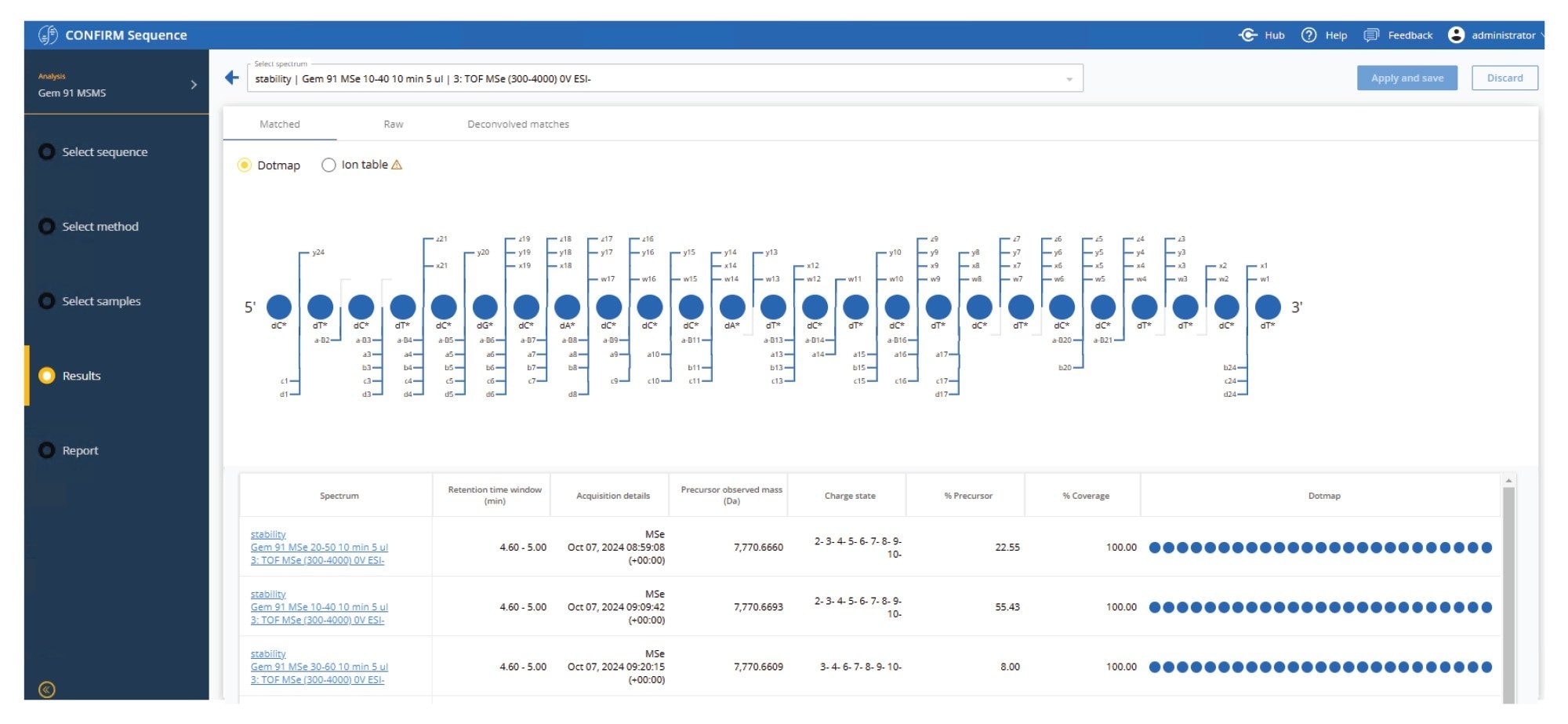
In complement, the waters_connect CONFIRM Sequence app, can be used to facilitate processing of data-independent MSE fragmentation spectra for oligonucleotide sequence verification and localizing sites of modification. The sequence coverage can be viewed in a "dot-map" visualization to represent the sequence coverage for both full-length products and lower abundance oligonucleotide impurity modifications. This functionality is crucial for the rapid and efficient interpretation of complex oligonucleotide fragmentation spectra, supporting the growing demands of oligonucleotide therapeutic pipelines, and the pressures exerted on development, manufacturing and quality control organizations to make faster and more confident decisions.2
By providing these detailed insights into the molecular composition of oligonucleotides, the Xevo MRT MS helps ensure consistency, stability, and quality of the final product, making it an invaluable tool for not only characterization, but also for continually evaluating the quality and consistency of biotherapeutic products and processes.
Experimental
Sample
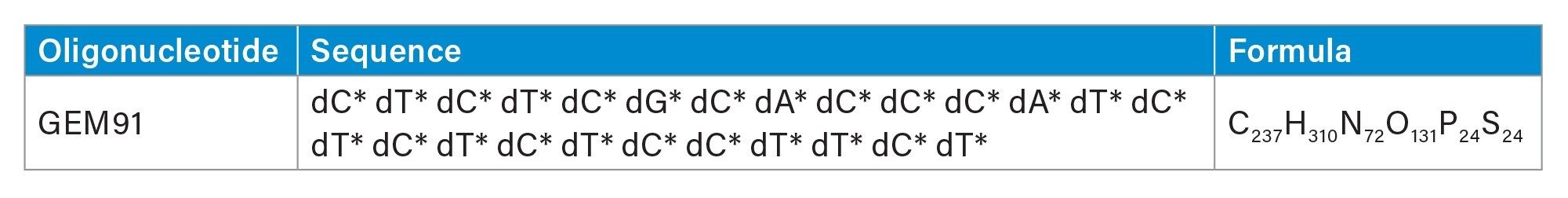
A well characterized 25-mer phosphorothioate antisense oligonucleotide (GEM 91, Trecovirsen) was analyzed using ion pair reversed phase IPRP-LC-MS in negative ESI mode.
Triethylamine (TEA, 99.5% purity, catalogue number 65897-50ML) and methanol (LC-MS grade, catalogue number 34966-1L) were obtained from Honeywell, Waters™ IonHance™ HFIP (1,1,1,3,3,3- hexafluoro-2-propanol p/n: 186010781-10ml) obtained from Waters and HPLC grade deionized (DI) water was purified using a MilliQ system. Mobile phases were prepared fresh and used on the same day.
LC Conditions
|
LC system: |
ACQUITY Premier UPLC System (Binary) |
|
Detection: |
UV 260 nm |
|
Vials: |
TruView™ LCMS Certified 12 x 32 mm Screw Neck Vial, Total Recovery, with Cap and Preslit PTFE/Silicone Septum (p/n: 186005663CV) |
|
Column: |
ACQUITY Premier Oligonucleotide C18 Column 130 Å, 1.7 µm 2.1 mm x 50 mm (p/n: 186009484) |
|
Column temperature: |
60.0 °C |
|
Sample temperature: |
6.0 °C |
|
Injection volume: |
5 µL |
|
Flow rate: |
0.400 ml/min |
|
Mobile phase A1: |
7 mM triethylamine (TEA) and 40 mM IonHance HFIP in Milli-Q water (pH 8.6) |
|
Mobile phase B1: |
3.5 mM TEA, 20 mM HFIP in 50% methanol |
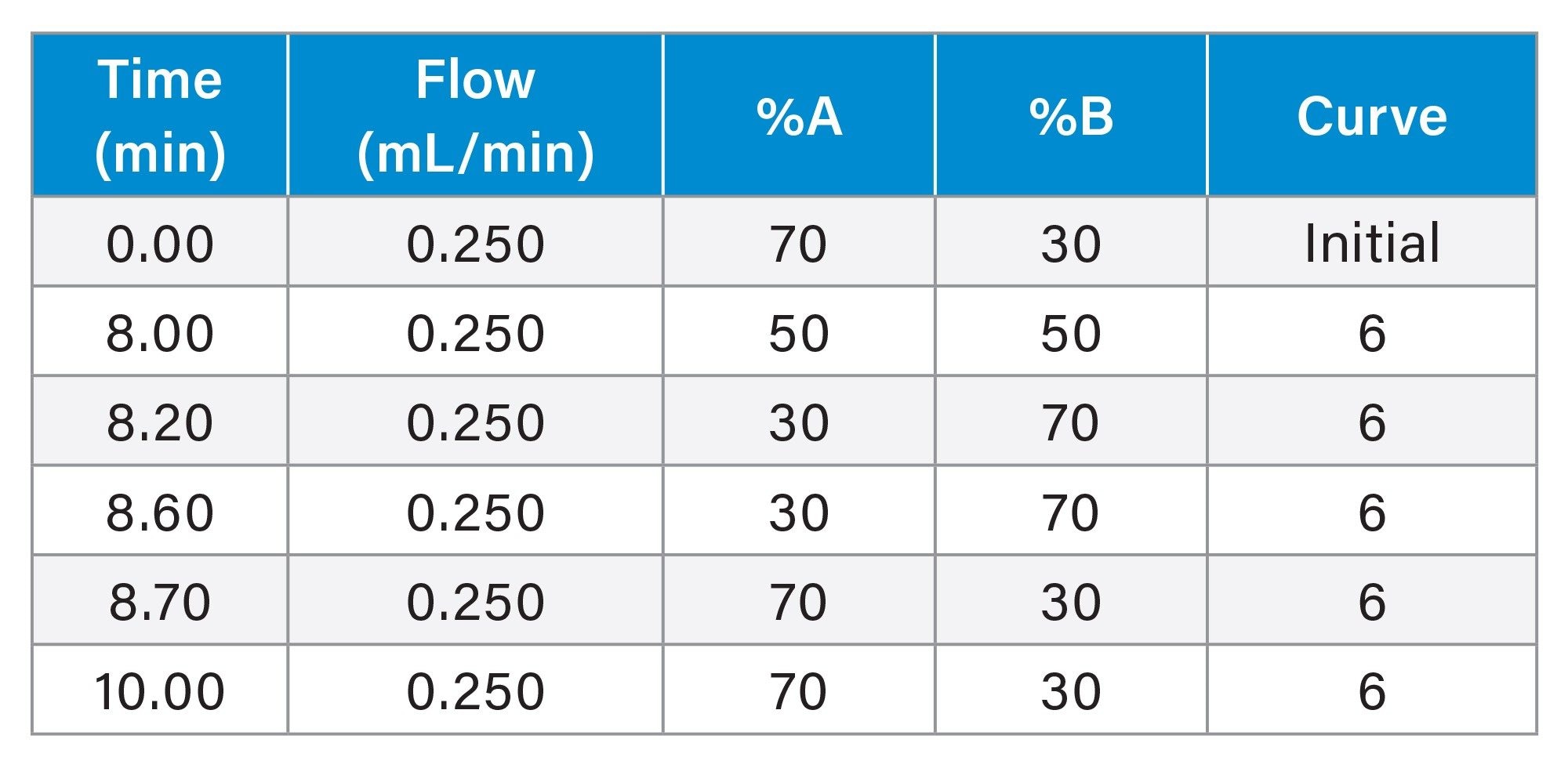
Gradient Table
MS Conditions
|
MS system: |
Xevo MRT MS |
|
Mode: |
MSE |
|
Mass range: |
400–4000 m/z |
|
Polarity: |
Negative |
|
Scan rate: |
10 Hz |
|
Cone voltage: |
40 V |
|
Cone Gas: |
50 L/hr |
|
Source temperature: |
120 °C |
|
Desolvation temperature: |
400 °C |
|
Desolvation gas: |
800 L/hr |
|
Capillary voltage: |
2.20 kV |
|
MSE Collision energy ramp: |
10–20 eV |
Results and Discussion
GEM 91 (Trecovirsen), a 25-mer phosphorothioate antisense oligonucleotide (ASO), was used for this study to assess the routine performance of the Xevo MRT MS System for confirming FLP identity, generating impurity profiles, and fragmentation-based impurity localization.
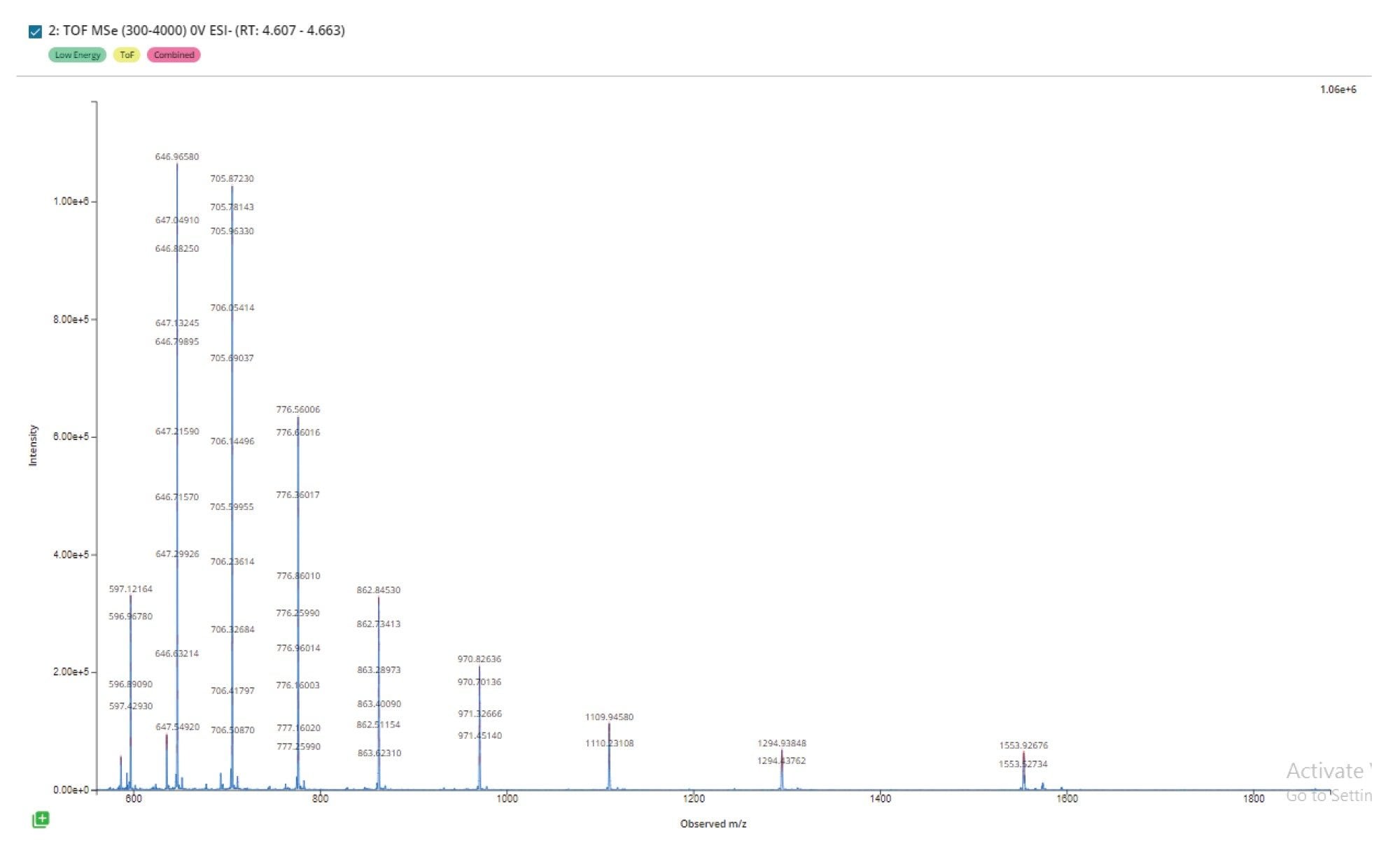
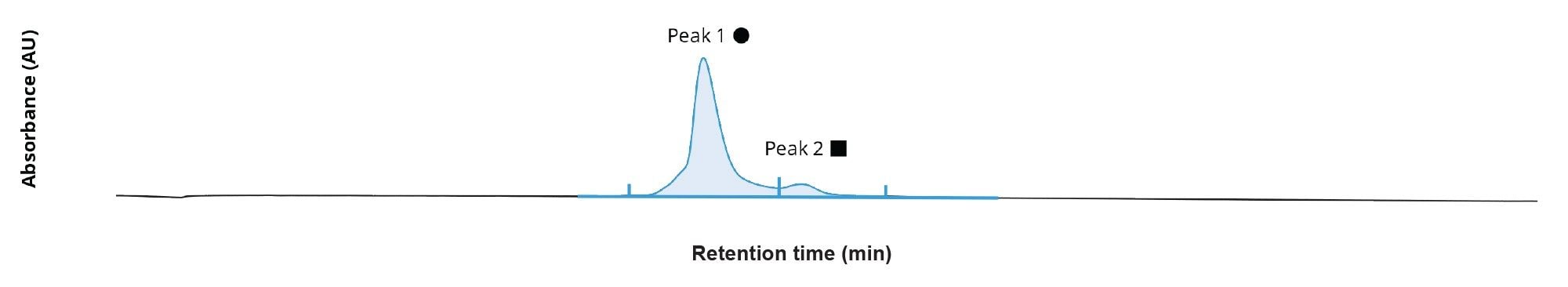
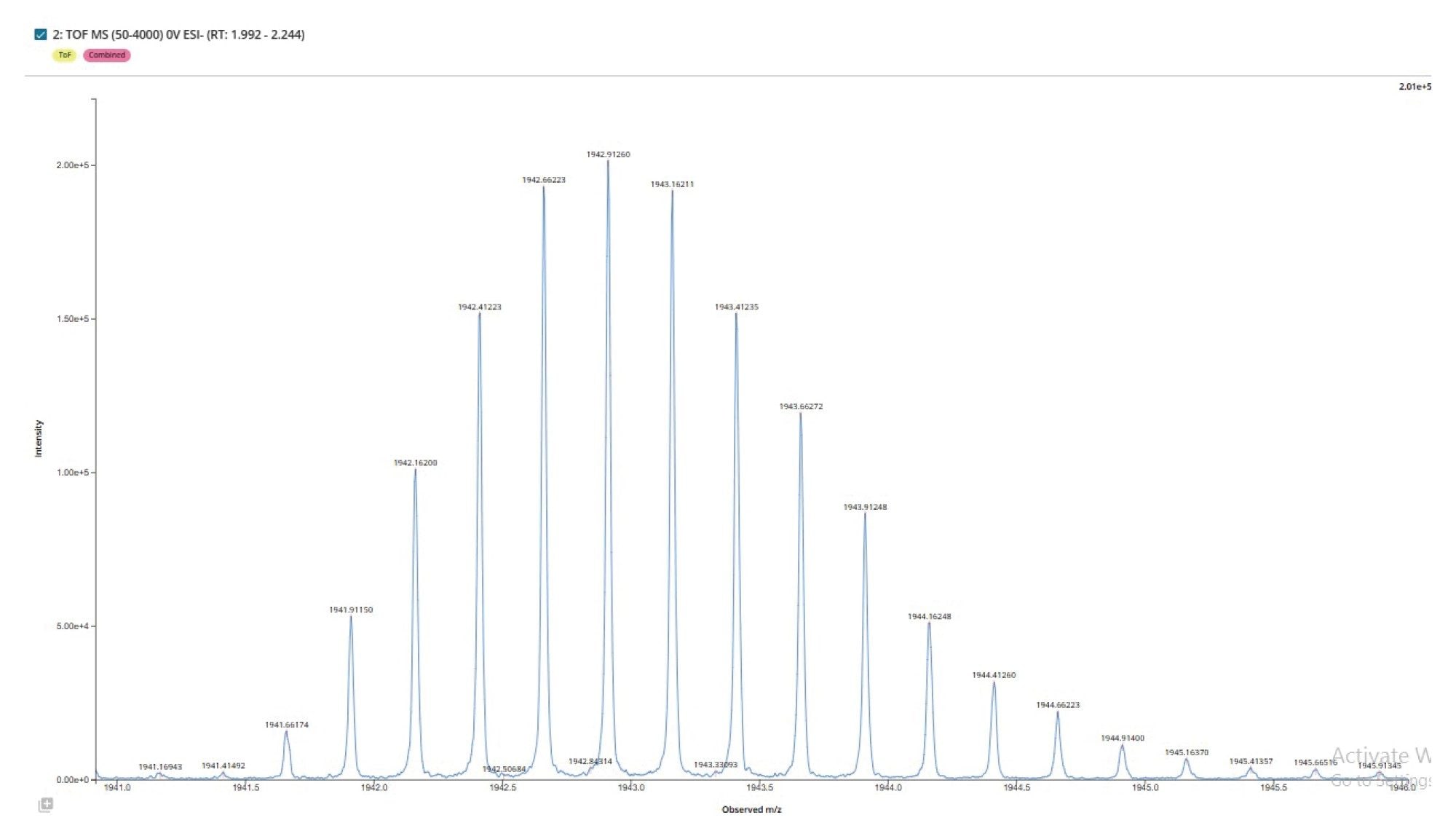
The automated detection of chromatographic peaks by the INTACT Mass App processing (Figure 2), enabled summed spectra representing the full charge state envelope to be produced for the GEM 91 full length product (Figure 3), with a closer view of the isotopic structure for the 4- charged state (Figure 4). The low level of solvent adducts combined with efficient desolvation, generated spectra with minimal depurination artifacts (~0.4%) or observable solvent adducts.
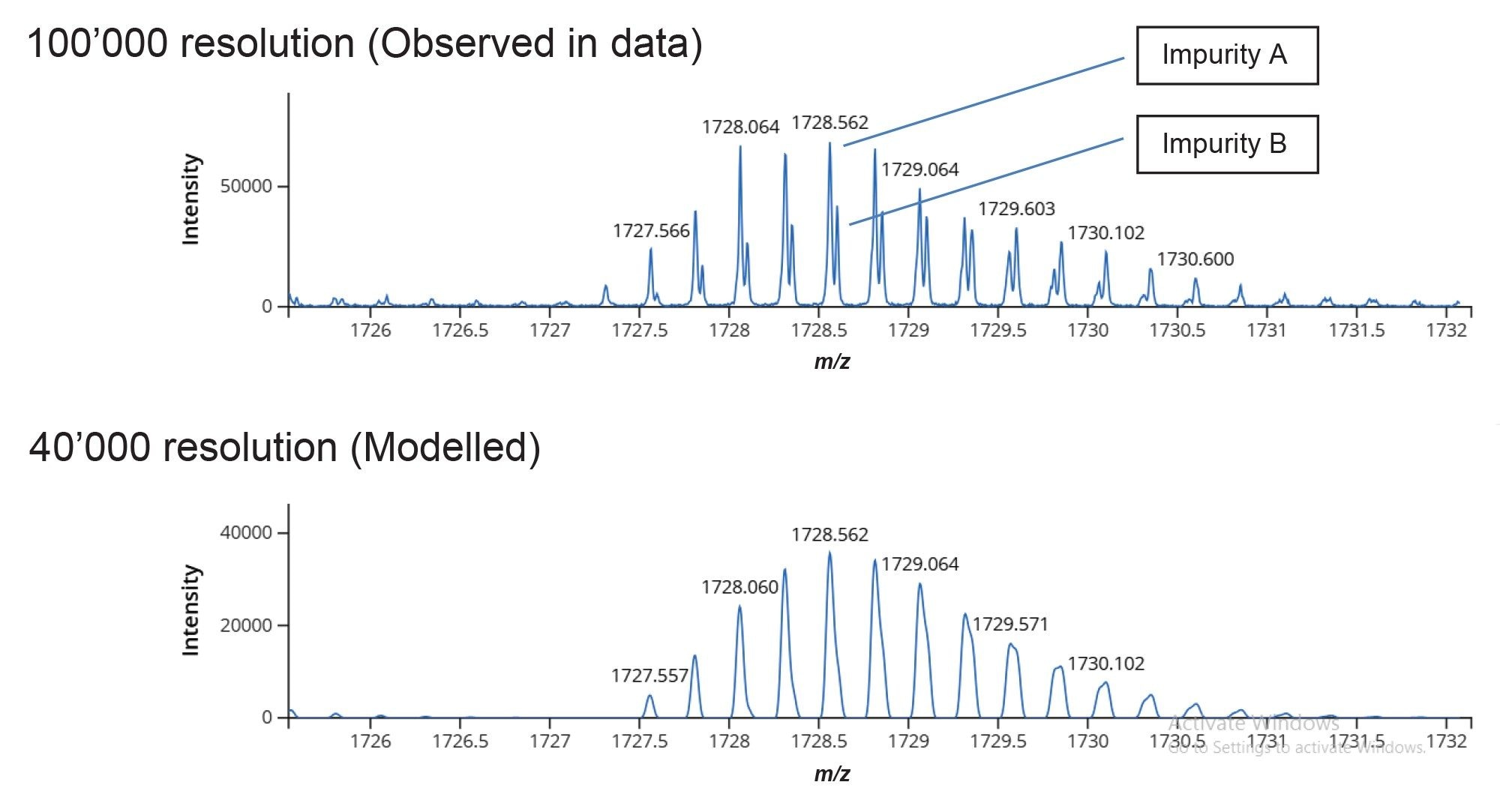
With a mass spectral resolution of approximately 100,000 FWHM in this data we could clearly distinguish two impurities at m/z 1728.56 and 1728.60 (Figure 5, Top), with less than 0.05 Da difference between the [M-4H]4- impurity ions. A theoretical model spectrum with a simulated resolution of 40,000 FWHM (Figure 5, Bottom) demonstrates how only a wider fused peak containing both species would be observed, highlighting the utility of high mass spectral resolution and isotopically resolved spectra for detecting coeluting impurities with small mass differences.
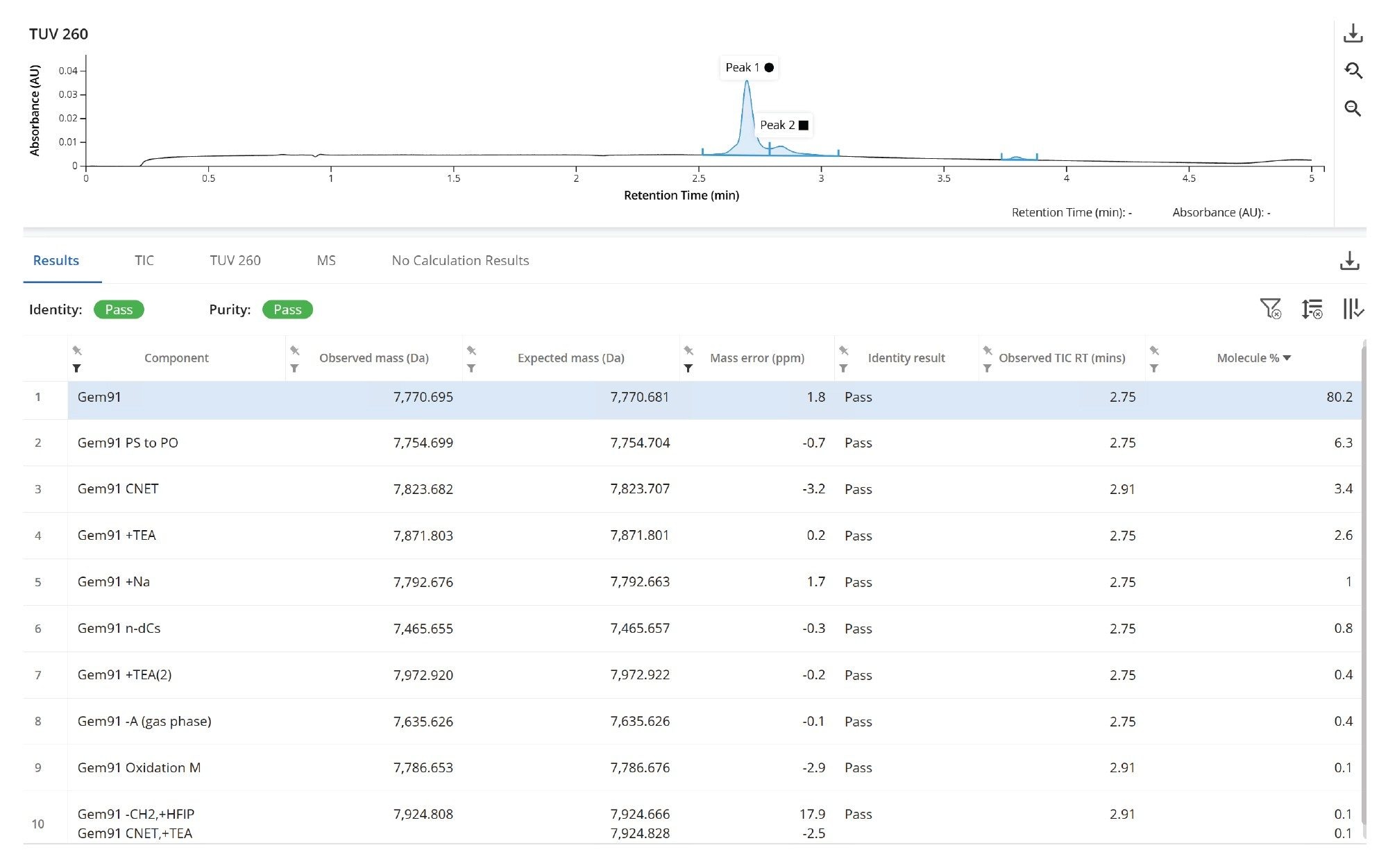
Automated processing (automated chromatographic peak detection and BayesSpray deconvolution of the summed spectra of the main peak) with the INTACT Mass App was used to confirm the mass of the full-length oligonucleotide product and assess the purity profile. INTACT Mass App deconvoluted spectral results for the GEM 91 sample matched the predicted mass of the full length product within 2 ppm, and revealed 9 potential impurities assigned to common synthetic impurities observed to the 0.1% level relative to FLP signal. Again, all the impurities were identified with high mass accuracy ranging from 3 ppm down to sub-ppm mass errors. This level of mass accuracy reduces the likelihood of misassignments, streamlining the data processing workflow and saving valuable time. For the 0.1% impurity observed at mass 7924.808 Da, there are two potential matches: one at –2.5 ppm and another at 17.5 ppm (Figure 6). Based on the observed mass accuracy of the FLP and other assigned modifications, we can confidently infer that the +CNET, +TEA combination would be the correct modification assignment.
The Xevo MRT MS utilizes a novel collision cell geometry and employs a Nitrogen collision gas for greater control over fragmentation, improving fragment ion transmission in both targeted MS/MS or MSE data independent acquisition (DIA) modes. Unlike traditional data-dependent acquisition methods, MSE employs data-independent acquisition, capturing MS2 fragment ions for all MS1 ions in a single run, ensuring consistent fragmentation data for all precursor ions in the MS1 data. This approach provides for high sequence coverage of the FLP and its’ impurities, supported by accurate mass fragmentation data, and sufficient MS1 data points to enable precursor ion peak quantification, all within a single injection.
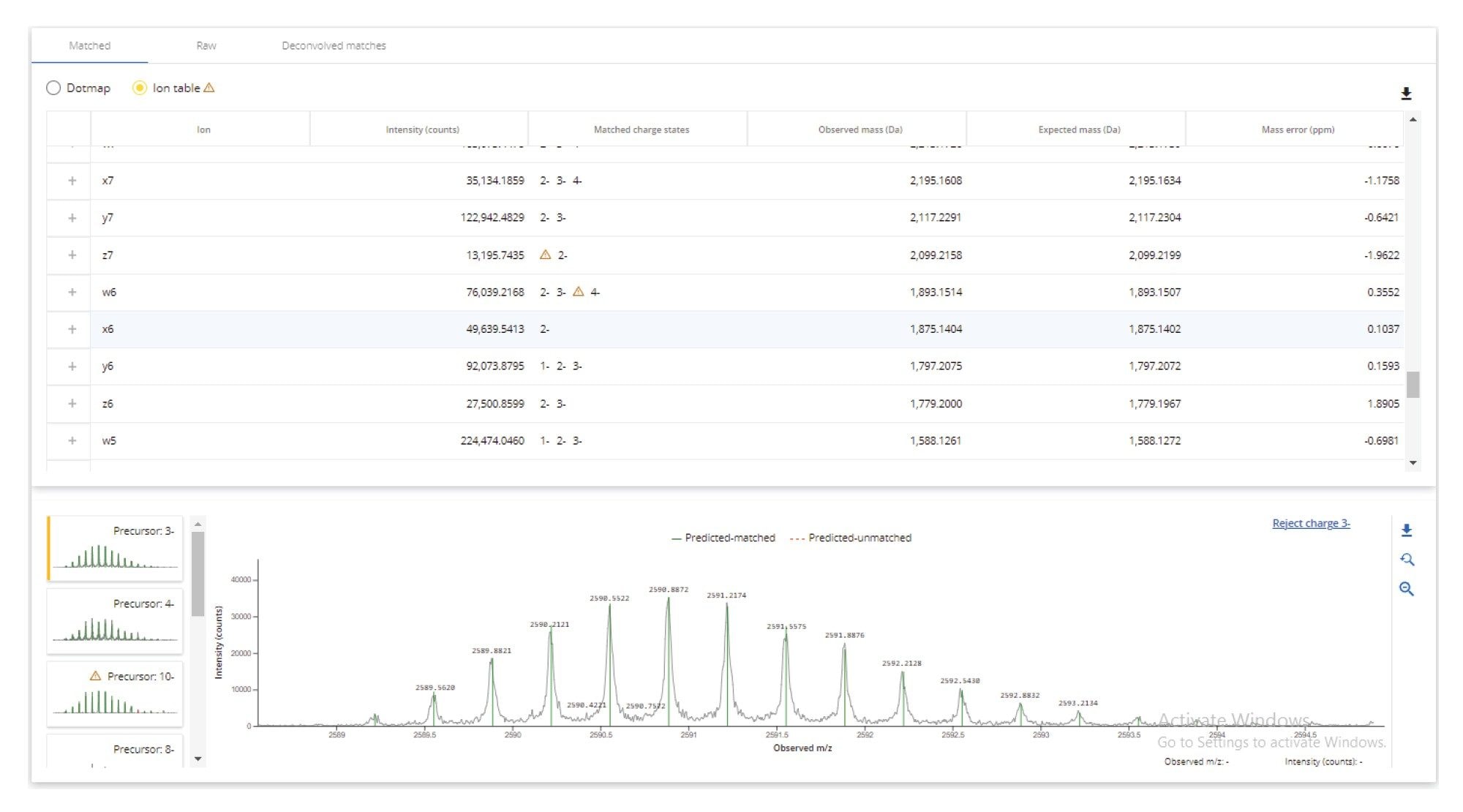
The collected GEM 91 MSE data were processed using the waters_connect CONFIRM Sequence App to verify sequence against the in-silico predicted GEM 91 fragmentation spectra. Fragmentation coverage can be viewed in a "dot-map" format (Figure 7), which allows for quick confirmation of the predicted FLP sequence and of impurities predicted by the intact oligo profile. In this case, 100% fragmentation coverage was achieved for the GEM 91 FLP (Figure 8) with fragment ion assignment mass accuracies of under 2 ppm, demonstrating high precision mass measurement of both precursor and fragment ions.
Conclusion
With the dramatic advancement of pipelines for oligonucleotide-based therapeutics, the pressure to make quick, high-quality decisions for product and process development has never been higher. The Xevo MRT MS System offers numerous benefits for detailed oligonucleotide characterization and for continued monitoring of product variation as a molecule progresses through development and commercialization. High sensitivity, dynamic range and sub- to low-ppm mass accuracy enables precise determination of oligonucleotide mass, sequence confirmation for product identity and assessment of lower abundance modifications and impurities for increasingly complex oligonucleotides.
This exceptional MS detection performance of the Xevo MRT MS was complemented with a streamlined, automated, compliant-ready informatics workflows within the waters_connect INTACT Mass and CONFIRM Sequence Applications that supported integrated data acquisition, processing, review, and reporting for Oligonucleotide data acquired from individual samples.
Overall, the Xevo MRT MS System under waters_connect informatics has been demonstrated to be a robust and efficient platform for oligonucleotide analysis, enabling biopharmaceutical analysts to make confident development and commercialization decisions without the complications that can arise from manual data processing and reporting using spreadsheets and other 3rd party software tools.
For more information, please visit https://www.waters.com/nextgen/global/products/mass-spectrometry/mass-spectrometry-systems/xevo-mrt.html
References
- Intact Mass Confirmation Analysis on the BioAccord LC-MS System for a Variety of Extensively Modified Oligonucleotides.
- A Xevo™ G3-based Workflow for Purity Determination, Intact Mass Measurement, and MS/MS Sequencing of Impurities Detected in Synthetic Oligonucleotides.
- an-automated-compliance-ready-lc-ms-workflow-for-intact-mass-confirmation-and-purity-analysis-of-oligonucleotides.html.
720008764, April 2025