
This is an Application Brief and does not contain a detailed Experimental section.
For research use only. Not for use in diagnostic procedures.
The COVID-19 outbreak has prompted the development of LC-MS based methods to detect viral infection and load as a complement to current tests to determine infection. Targeted mass spectrometry, through the detection of viral proteins in proteolytically digested body fluids, has been suggested as an additional SARS-CoV-2 test method. The work presented here demonstrates the application of UniSpray and Electrospray ionization sources for the detection of selected SARS-CoV-2 tryptic peptides with a Xevo TQ-XS Tandem Quadrupole Mass Spectrometer.
COVID-19 coronavirus disease is a highly pathogenic viral infection caused by SARS-CoV-2 and responsible for the current on-going pandemic. SARS-CoV-2 virus particles are protein rich, with Spike glycoprotein (SPIKE) and Nucleoprotein (NCAP) being the two main constituents. The detection and quantification of SARS-CoV-2 proteins by a targeted LC-MS method is being considered as an alternative technology for COVID-19 viral load determination to complement polymerase chain reaction (PCR) based testing.1,2 Therefore, a community-based effort to develop a ‘A Universally Adoptable Corona Multiple Reaction Monitoring Assay’ was initiated.3 Here, we have compared a novel ionization technique UniSpray with Electrospray ionization to further optimize the LC-MS detection method (720006967 and 720006968) for selected SPIKE and NCAP tryptic peptides and investigate the potential increase in dynamic range and selectivity of this LC-MS method.
Tryptic-Lys C peptides from a combined digestion procedure of recombinant SARS-CoV-2 SPIKE and NCAP proteins, as individual standards and spiked in Universal Transport Medium (UTM) matrix, respectively, were obtained in freeze-dried form from Cov-MS.3 The resulting peptides were analyzed in MRM mode of analysis using an ACQUITY UPLC I-Class PLUS System interfaced to a Xevo TQ-XS Tandem Quadrupole Mass Spectrometer.
|
LC system: |
ACQUITY UPLC I-Class PLUS |
|
Vials: |
QuanRecovery Vials with MaxPeak HPS |
|
Column(s): |
ACQUITY Premier Peptide BEH C18 300 Å, 2.1 mm x 50 mm, 1.7 µm |
|
Column temp.: |
40 °C |
|
Sample temp.: |
10 °C |
|
Injection volume: |
5 µL |
|
Flow rate: |
0.6 mL/min |
|
Mobile phase A: |
0.1% formic acid in H2O |
|
Mobile phase B: |
0.1% formic acid in acetonitrile |
|
MS system: |
Xevo TQ-XS |
|
Ionization mode: |
UniSpray and ESI positive |
|
Acquisition mode: |
MRM |
|
Capillary voltage (ESI+): |
0.5 kV |
|
Impactor voltage (US+): |
0.7 kV |
|
Collision energy: |
peptide/transition optimized |
|
Cone voltage: |
35 V |
|
Time (min) |
%B solvent |
|---|---|
|
0.0 |
5 |
|
5.5 |
33 |
|
5.6 |
85 |
|
7.0 |
85 |
|
7.1 |
5 |
|
8.0 |
5 |
|
Software: |
MassLynx TargetLynx |
The developed Multiple Reaction Monitoring (MRM) method described in Comprehending COVID-19: Maximizing LC-MS Detection Dynamic Range for Multiple Reaction Monitoring Based SARS-CoV-2 Analysis (720006968) as part of a community based effort in collaboration with the Cov-MS consortium was applied in this study as well.3 It uses two transitions per peptide in order to maximize duty cycle and signal-to-noise, as well as regular LC methods and conditions in order to maintain robustness and throughput. Both UniSpray and Electrospray ionization interfaces were applied, and the same dilution series samples analyzed as described in the original Cov-MS standard operation procedure (SOP).
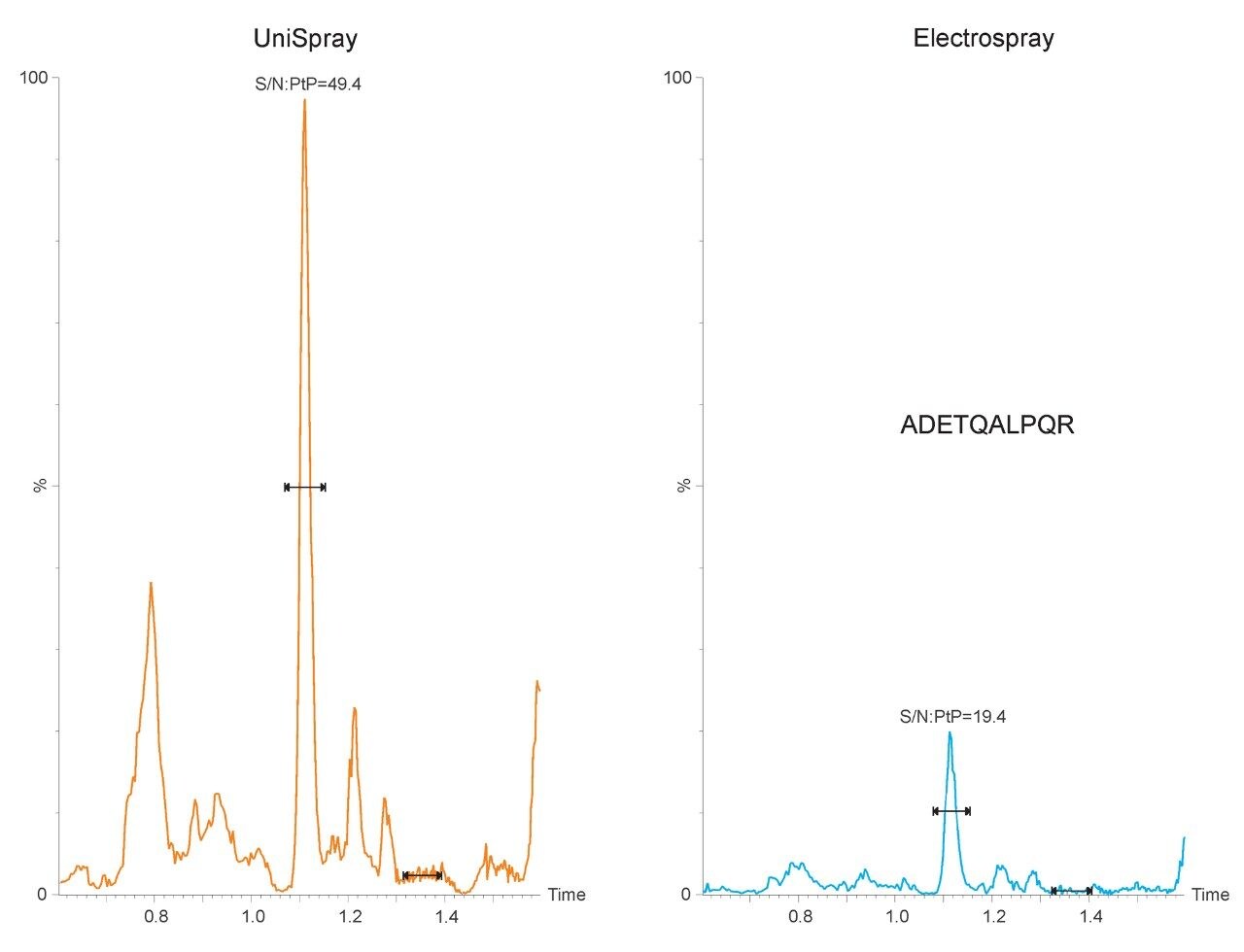
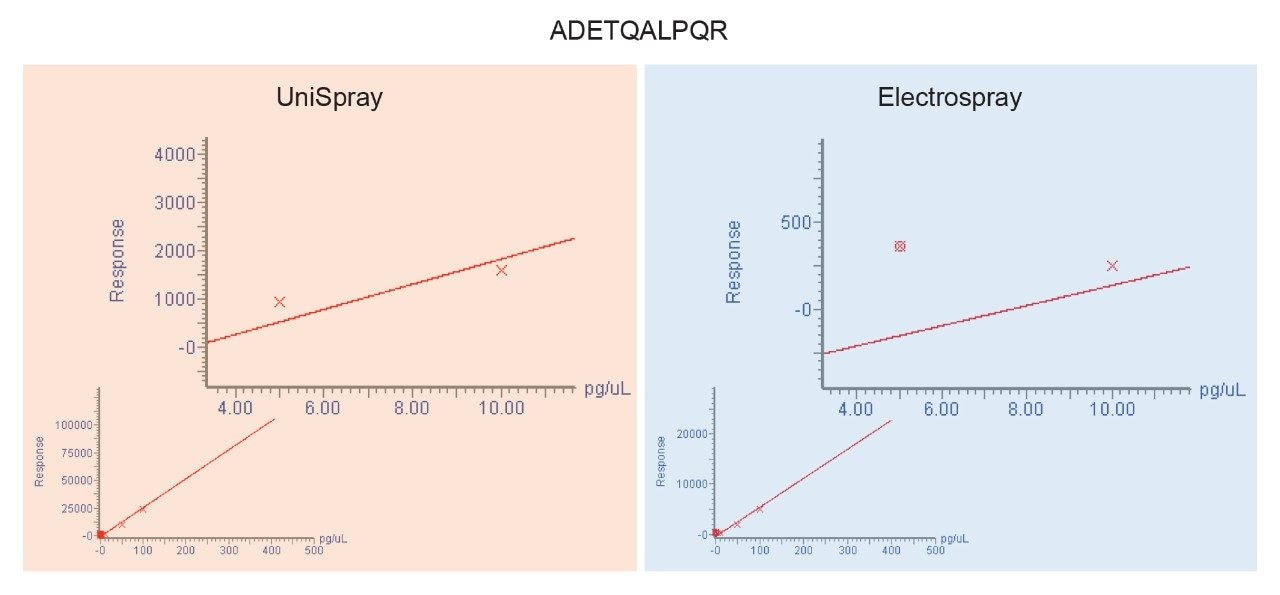
The chromatograms shown in Figure 1 are typical in that the application of UniSpray results in higher signal intensities compared to Electrospray. However, in this example, UniSpray also provided improved S/N compared to its Electrospray counterpart. The S/N gain obtained for this peptide at this amount of protein digest injected on-column was approximately two-and-a-half, which is in agreement with previously reported performance metrics for peptides.4 The benefit of improved S/N is shown in Figure 2, where the quantitative response of peptide ADETQALPQR from P0DTC9|NCAP_SARS2 obtained with UniSpray and Electrospray interfaces is demonstrated. Peptide ADETQALPQR was previously identified as one of the medium-to-better responding peptides, covering four amounts levels injected on-column. With this peptide and matrix, using UniSpray, an additional amount level could be reached without compromising quantitative performance in terms of residuals and linear regression.
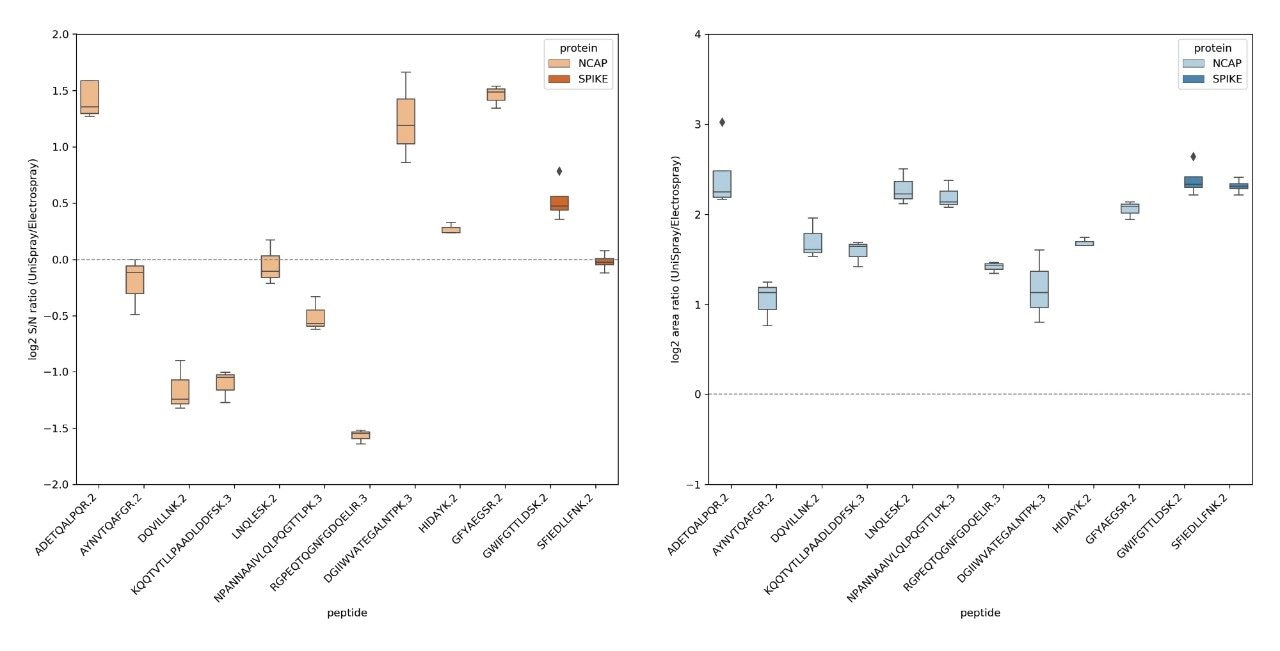
A comparative results summary is provided in Figure 3, contrasting the peak S/N and area of all MRM chromatograms for the peptides from the NCAP and SPIKE SARS-Cov-2 proteins specified in the Cov-MS SOP, illustrating the complementary nature of the UniSpray and Electrospray interfaces. An average S/N ratio and range was calculated for each peptide by expressing S/N for a given peptide at a particular amount level and contrasting it against the S/N for the same peptide at the same amount level. Next, an average and confidence interval of all S/N ratio values for all amount levels at which the peptide was detected was expressed as illustrated by the quartile distributions shown in the left-hand side set of ratio values of Figure 3. On average, about three peptides showed improved S/N using UniSpray and a similar number of peptides showing improved S/N using Electrospray. However, the ability to have the option to select or use an alternative technology can be critical in being able to detect more confidently particular SARS-Cov-2 peptides in a given matrix, i.e. nasopharyngeal swabs, saliva, gargle solution, etc. The right-hand side set of distributions, shown in Figure 3, demonstrate the peak area ratio for both ionization techniques, confirming the observation illustrated in Figure 2 and previous studies that the application of the UniSpray interface provided overall larger peak areas, i.e. an average 4-fold increase compared to Electrospray, providing better ion statistics, thus potentially improved reproducibility at lower data signal levels (data not shown).4
Since the NCAP and SPIKE proteins are key components of the viral SARS-CoV-2 complement and a direct measure of viral load, LC-MS based technologies are being considered to determine their amounts and concentration in biological matrices. Here, the linear response and LLOD of a number of tryptic NCAP and SPIKE peptides detected in UTM matrix have been investigated to understand the relative response of UniSpray and Electrospray ionization sources and to further characterize the analytical MRM method under development by the Cov-MS consortium. The obtained results indicate that UniSpray and Electrospray are complementary ionization methods for proteolytic peptides and that the developed Xevo TQ-XS Tandem Quadrupole Mass Spectrometer method to detect and quantify SARS-CoV-2 proteins in nasopharyngeal swabs and preserved in Universal Transport Medium could potentially be further enhanced using a subset of peptides.
The Cov-MS consortium is kindly acknowledged for making evaluation kits available as part of a community-based effort to design a SARS-Cov-2 MRM method.
Stuart Oehrle, Laurence Van Oudenhove, Jan Claereboudt, and Hans Vissers (Waters Corporation); Bart Van Puyvelde, Simon Daled, Dieter Deforce, and Maarten Dhaenens (Pharmaceutical Biotechnology, University of Ghent); Katleen Van Uytfanghe, (Department of Bioanalysis, University of Ghent).
720007054, September 2020