For research use only. Not for use in diagnostic procedures.
In this study, instrumental and application benefits are demonstrated for the identification of proteins and peptides in a new high definition data directed analysis (HD-DDA) mode, where ion mobility spectrometry is incorporated into a quadrupole time-of-flight mass spectrometer. HD-DDA uses a high duty cycle mode and enhanced decision making to provide a highly sensitive and selective experiment.
***nanoACQUITY UPLC applications readily transfer to the ACQUITY UPLC M-Class System***
The increasing complexity of bottom-up proteomics challenges the capabilities of mass spectrometers to generate more and more detailed information. Instrument speed, sensitivity, and mass accuracy have all increased significantly over recent years, thereby affording better quality data, improved peptide sequence annotation, and more accurate identification results.
In line with improved hardware features, novel LC-MS acquisition schemas and fragmentation mechanisms have been introduced, including parent ion discovery (PID) methods, data independent acquisitions (DIA), ion mobility (IM) assisted methods, and electron transfer dissociation (ETD). To date, IM has been mainly employed for the cross sectional and structural analysis of various analyte types,1 and enhancing the specificity of DIA acquisitions such as HDMSE.2
In this study, instrumental and application benefits are demonstrated for the identification of proteins and peptides in a new high definition data directed analysis (HD-DDA) mode, where ion mobility spectrometry is incorporated into a quadrupole time-of-flight mass spectrometer. HD-DDA uses a high duty cycle mode and enhanced decision making to provide a highly sensitive and selective experiment.
The cytosolic content of E.coli and hela cells were digested using trypsin. Lysates were injected on a nanoACQUITY UPLC System equipped with an ACQUITY UPLC BEH 1.7 μm, 15 cm x 75 μm column coupled to a SYNAPT G2-Si Mass Spectrometer. Data were processed and searched with ProteinLynx Global SERVER and/or Mascot.3
HD-DDA enhancements include full support for Wideband Enhancement,4 which affords a signal increase of five- to ten-fold as well as enhanced decision making logic when switching between MS and MS/MS modes. Wideband Enhancement utilizes ion mobility separation of product ions of a single charge state in combination with pusher synchronization to achieve nearly 100% duty cycle, as shown in Figure 1.
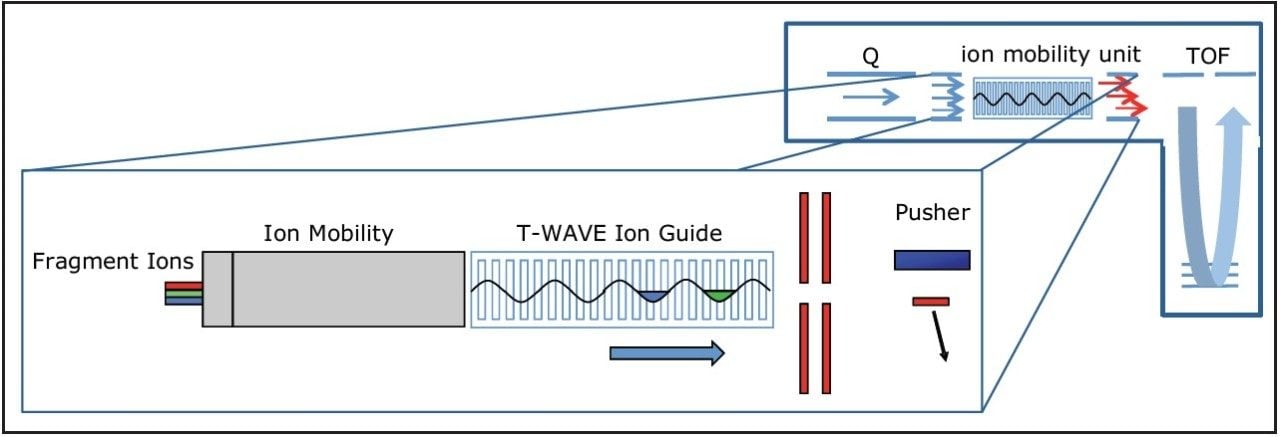
HD-DDA acquisitions are typically performed in an untargeted mode and can be complemented with unlimited include and/or exclude lists. Collision energies are stepped, ramped, or determined in real-time based on m/z and charge state. Data can be processed and searched with either ProteinLynx Global SERVER or vendor-neutral search algorithms and validation tools such as Mascot and Scaffold,5 respectively.
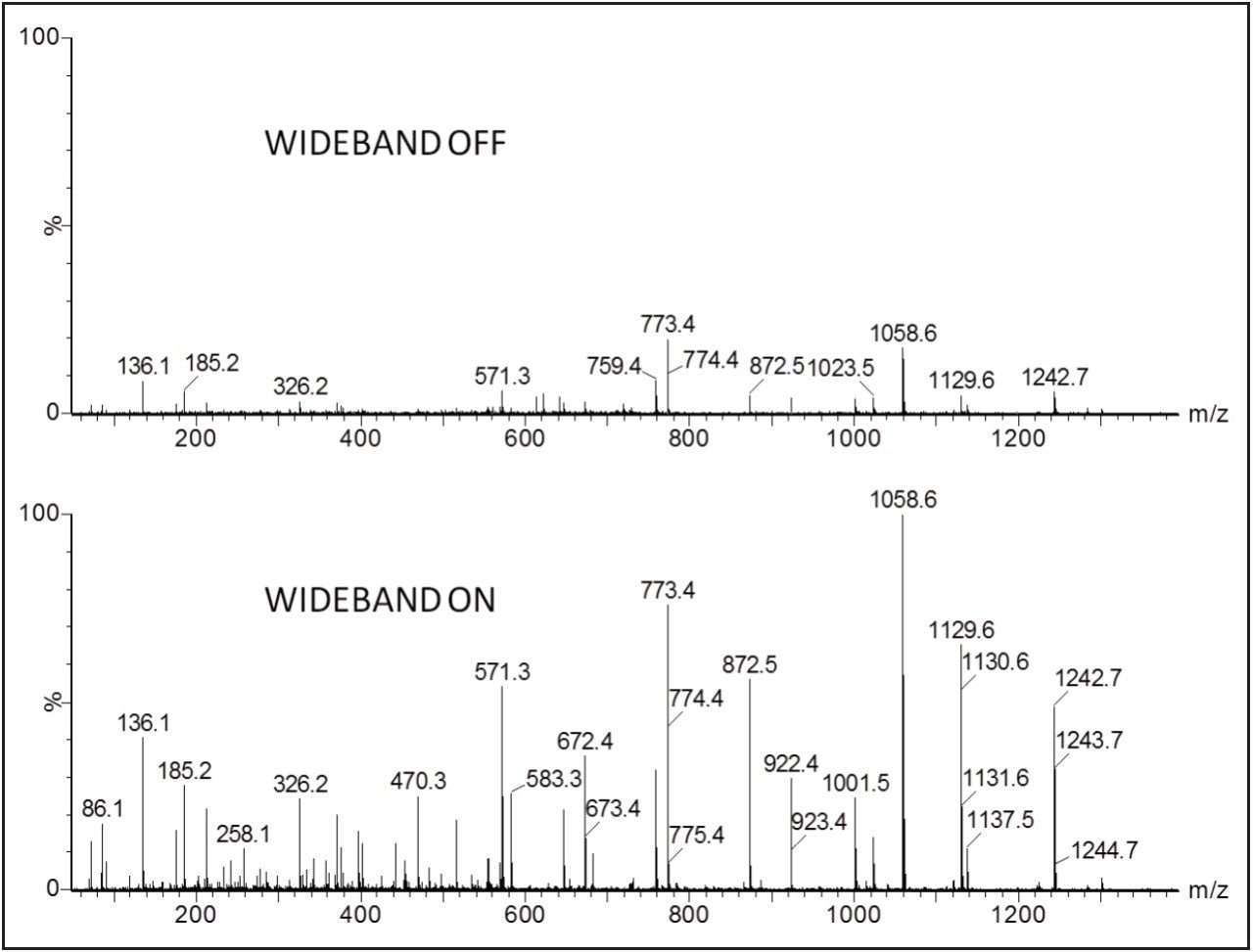
The benefit of Wideband Enhancement is demonstrated in Figure 2. Here, an E.coli tryptic digest was analyzed by means of nanoscale LC-MS/MS. Data were acquired by normal DDA and HD-DDA. In both cases, 0.1 seconds of MS/MS data were taken at the same moment of time within the LC peak. For this particular experiment, on average across the complete MS/MS spectrum, a five-fold signal increase was observed.
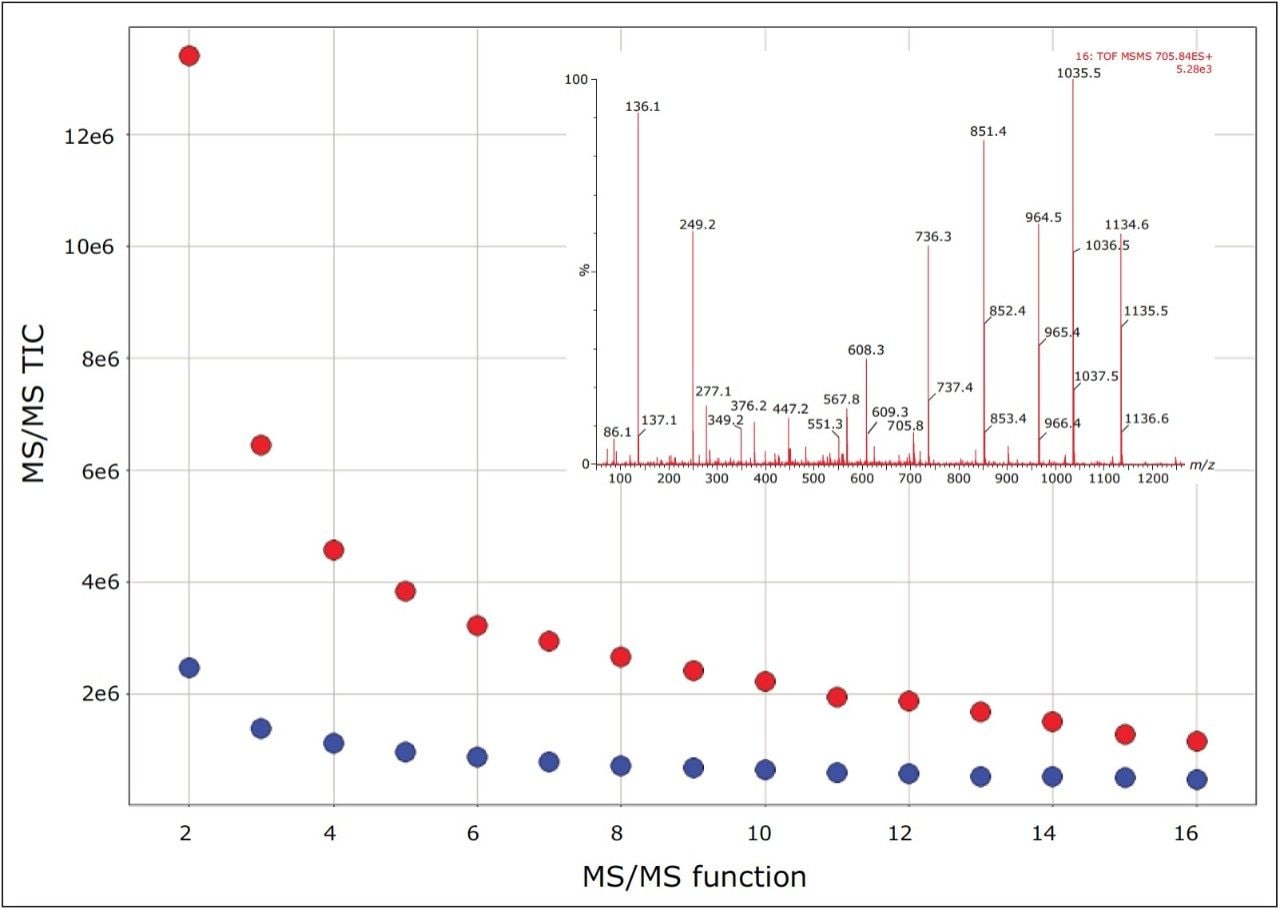
For this particular experiment, 15 concurrent MS/MS experiments were conducted per survey scan. The results, shown in Figure 3, illustrate the increase in MS/MS total ion current (TIC) as a function of the MS/MS channel when contrasting DDA with HD-DDA. The average increase per function was 420%, which is consistent with the results shown in Figure 2. The inset is an example MS/MS from the 15th channel, illustrating that MS/MS data with good signal-to-noise can be readily obtained from the lower abundant peptides present within the sample.
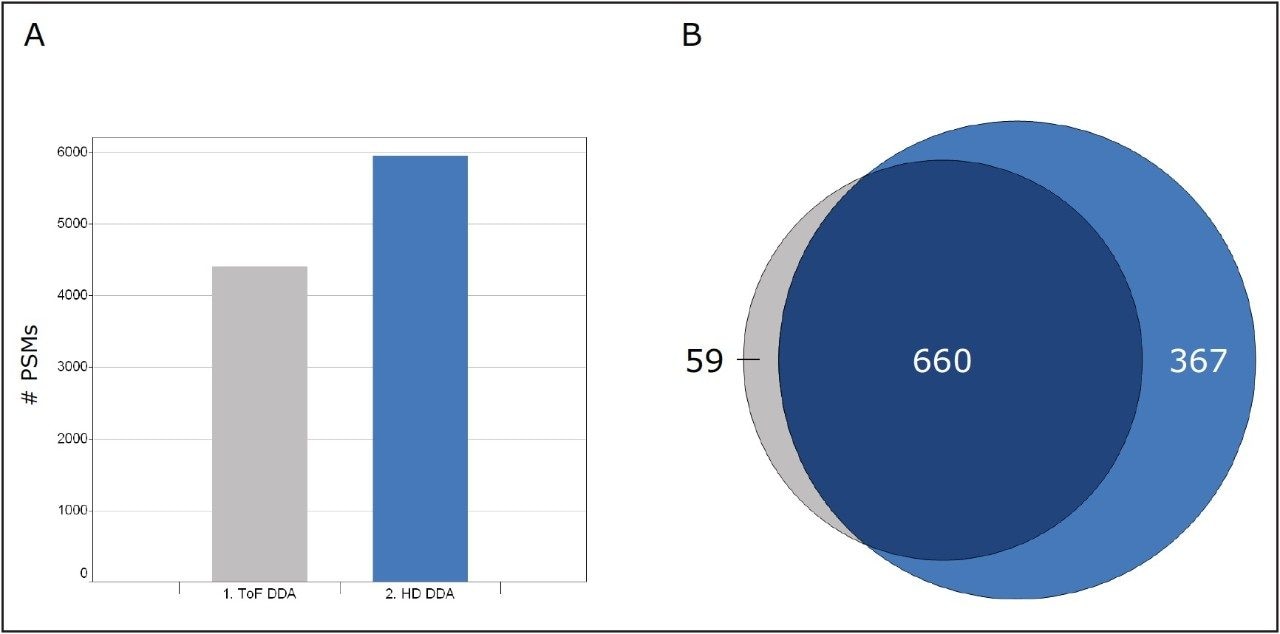
The increased sensitivity benefits afforded by HD-DDA for a bottom-up LC-MS proteomics experiment for the same E.coli sample are shown in Figure 4. Panel A shows the increase in number of peptides sequence matches. The Venn intersection in panel B contrasts the protein identifications. A significant increase in number of identified peptides (34.8%) and proteins (42.8%) was observed.
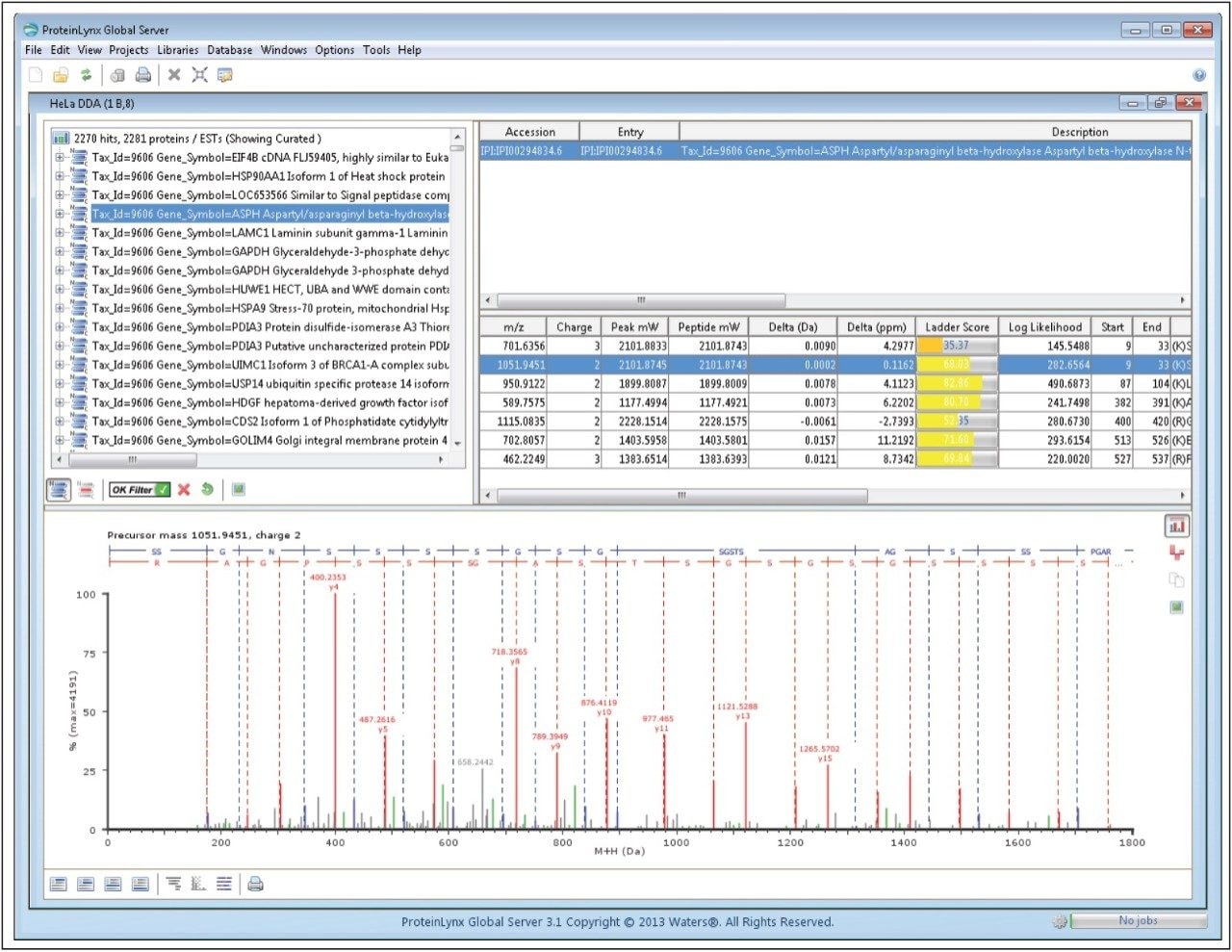
The search results from a more challenging sample are shown in Figure 5. Here, the PLGS search results are summarized for the analysis of a HeLA tryptic digest. A total of more than 2200 proteins were identified that passed a 95% identification confidence threshold. In this particular experiment, the spectrum identification rate was equal to 38%.
720004729, June 2013