
This application note demonstrates that the ACQUITY UPLC System configured with a tandem of 2.1 x 150 mm columns is capable of improving resolution of peptide maps and reducing coelution. This improved resolution is particularly useful for identification and quantification of protein heterogeneities with UPLC-UV.
LC-based peptide mapping is extensively employed for protein characterization, from early biotherapeutics development, to quality control of production and monitoring stability and activity after long-term storage. The use of this technology to identify and quantify low levels of impurity proteins, sequence variants, and post-translational modifications is critical for the assessment of protein drugs.
However, due to the complexity of protein enzymatic (often tryptic) digests, peptide coelution and poorly resolved chromatographic peaks are quite common in peptide mapping. In addition to recent advances in MS and particularly MSE,1 a good LC assay removes issues of ion suppression and isobaric interferences that may affect sensitive identification and accurate quantification of heterogeneities. Furthermore, a high-resolution LC assay can be more readily transferred for quality control purposes when utilizing UV detectors.
The use of sub-2-μm particles allows UPLC to push the limits of both peak capacity (higher efficiency) and sensitivity (sharper peaks). With UPLC, speed of analysis is also improved due to higher linear velocities.2 These features make UPLC-based peptide mapping powerful and attractive. Currently, the dimensions of 1.7-μm Peptide Separation Technology BEH C18 columns used with an ACQUITY UPLC System include 2.1 x 50 mm, 2.1 x 100 mm, and 2.1 x 150 mm. The pressure generated by these columns is far below the pressure limit (15,000 PSI) of the ACQUITY UPLC System at typical operating conditions (flow rate 0.2 mL/min, column temperature 20 to 65 °C), which leaves room for achieving further separation if longer columns are properly configured.
In this application note, we demonstrate how to maximize ACQUITY UPLC separation power by using a tandem of two 2.1 x 150 mm columns. The resolution of an IgG1 tryptic digest was compared between the tandem column configuration and single columns, with online detection by both a TUV detector and a SYNAPT MS system with MSE detection mode. The UPLC-UV/MSE reproducibility of the new configuration was evaluated. BiopharmaLynx 1.2 Software was used for identification of the eluted peptides.
|
LC System: |
ACQUITY UPLC with a standard peptide mapping mixer (425 μL) |
|
Column: |
Peptide Separation Technology (PST) BEH300 C18, 1.7 μm, 2.1 x 100 mm, 2.1 x 150 mm, or a tandem of two 2.1 x 150 mm columns |
|
Column temp.: |
65 °C |
|
Flow rate: |
200 μL/min |
|
Sample Injected: |
10 μL (100 pmole) |
|
Buffer A: |
0.02% TFA in water |
|
Buffer B: |
0.018% TFA in ACN |
|
Gradient: |
A linear gradient of 1-40% B was scaled with column lengths (100 mm, 150 mm, and 300 mm by coupling two 150 mm columns) for run times of 60, 90 and 180 min, respectively. |
|
Detection: |
TUV (214 nm) and MSE |
The SYNAPT MS System and MSE methods setup and operating conditions were the same as in previous descriptions.5-6
BiopharmaLynx 1.2 MassLynx Application Manager3,7
Waters MassPREP Enolase tryptic digestion standard was used. The IgG1 Antibody digest was prepared from a commercially available monoclonal antibody (mAb) by a RapiGest-assisted 4-h trypsin digestion protocol.3-4
A tandem of two 2.1 x 150 mm columns were coupled by capillary metal tubing and installed in the ACQUITY UPLC Column Manager. We first tested the system pressure generated by the configuration and the chromatographic reproducibility. At flow rate 200 μL/min and column temperature 65 °C, the system pressure was ≤7500 psi, far below the pressure limits of both the ACQUITY UPLC System (15,000 psi) and the 425 μL standard peptide mapping mixer (10,000 psi). We also tested the system pressure at different column temperatures, and demonstrated that the configuration with a tandem of two columns was feasible as a working system if the column temperature set above 40 °C.
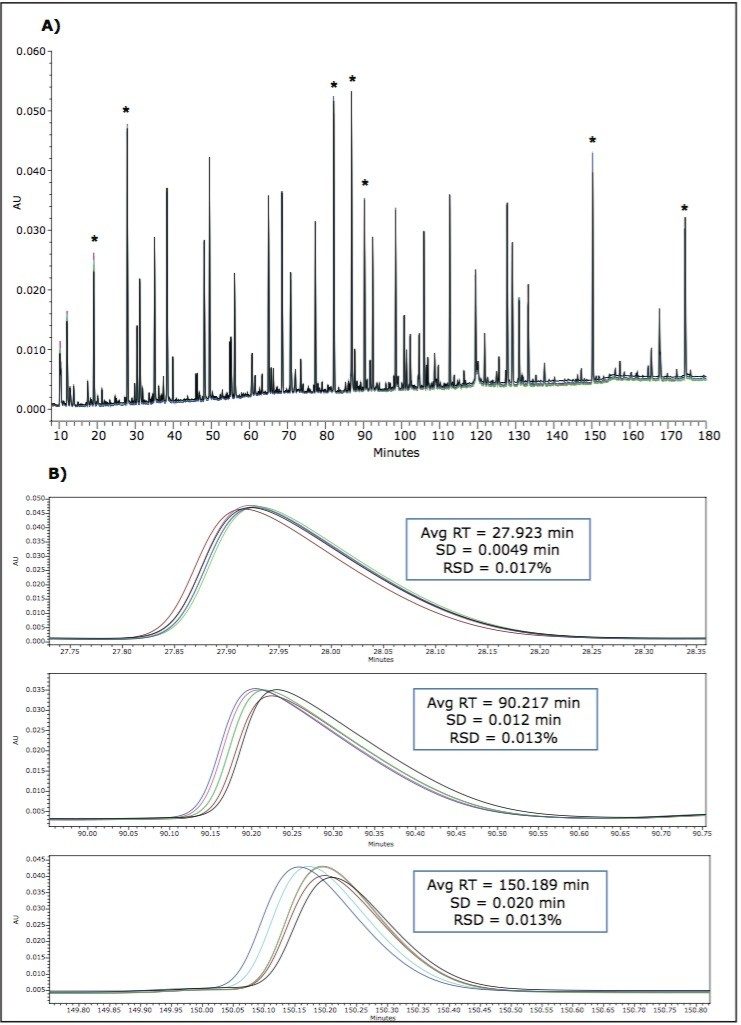
Excellent chromatographic repeatability was observed (Figure 1) in six continuous runs of MassPREP Enolase tryptic digestion standard using a gradient of 1-40% buffer B in 180 minutes. The retention time (RT) fluctuation from injection to injection is within 4 seconds for each eluted peak in the chromatograms. For example, the RT fluctuation observed for the lately eluted enolase tryptic peptide T37 (YPIVSIEDPFAEDDWEAWSHFFK, MW 2827.3 Da, RT around 150.19 min) is 3.3 seconds. The average relative standard deviation (RSD, calculated by standard deviation SD / average RT * 100%) is 0.017%. This proved the configuration did not affect the good reproducibility of the ACCQUITY UPLC System, as has been demonstrated in previous work studying ACCQUITY UPLC system-to-system reproducibility for peptide mapping.8
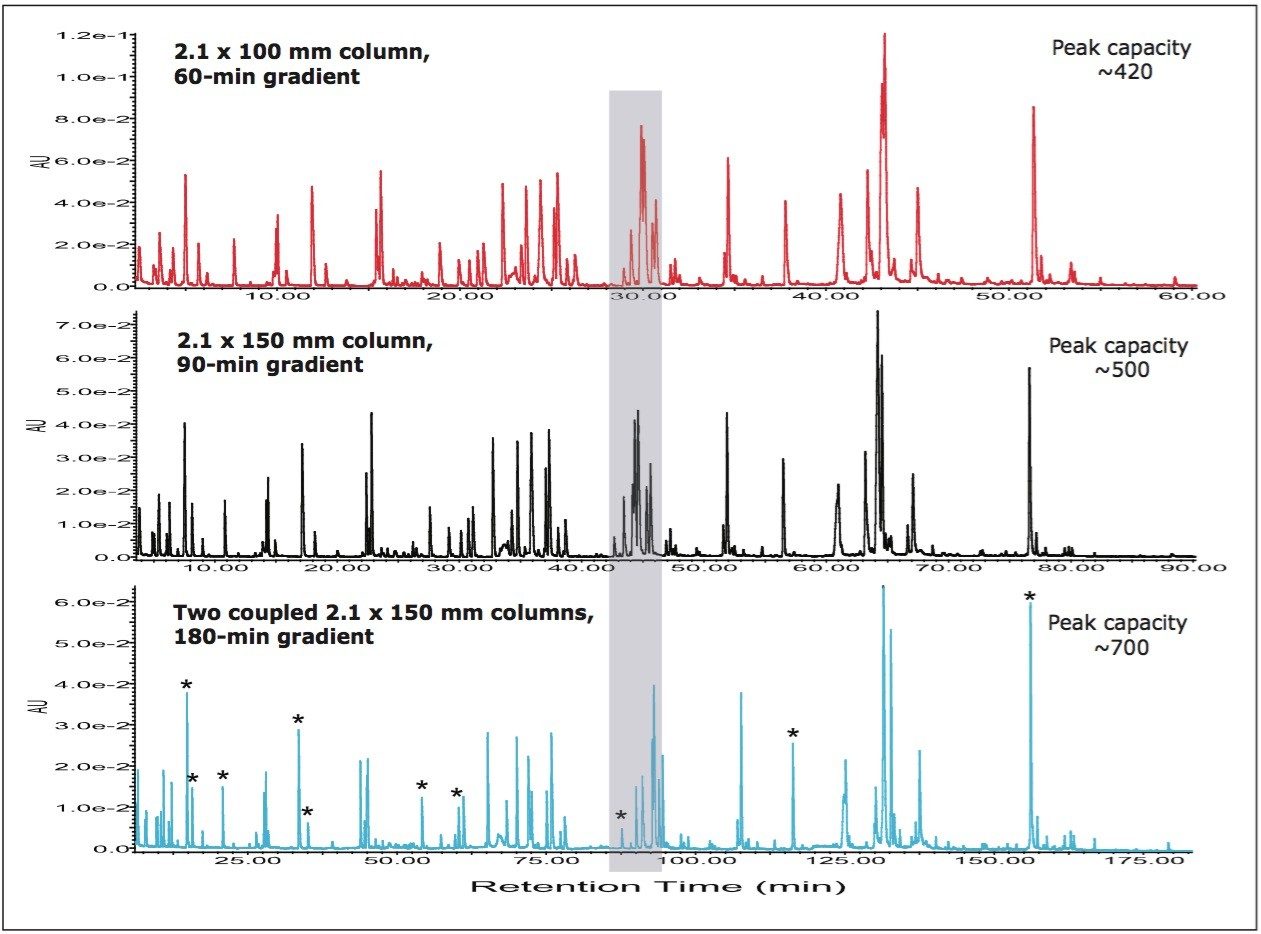
In order to evaluate the performance of the new configuration, we next compared the peptide maps of the IgG1 tryptic digest obtained between the tandem of two 2.1 x 150 mm columns and a single 2.1 x 100 or 2.1 x 150 mm column. The comparison was performed at same gradient slope by using a linear gradient of 1-40% B in run times (60, 90, and 180 min, respectively) scaled with column lengths (100 mm, 150 mm, and 300 mm of the tandem of two 150 mm columns). A good scalability and an apparently better separation were observed for the tandem columns (Figure 2). It clearly shows that poor or non-chromatographically-resolved peaks by single columns are separated by the tandem columns, e.g., those peaks in the region marked in shadow, due to significantly improved peak capacity. The calculated peak capacities for the 100 mm, 150 mm, and the tandem of two 150 mm columns in this peptide mapping experiment are about 420, 500, and 700, respectively.
Figures 3 and 4 further demonstrate the improved resolution of the tandem column configuration compared to the single columns by a zoom view of two detailed separation examples. The heavy chain tryptic peptides HT6 (IYDTNGYTR) and HT11* (AEDTAVYYC*SR, C* - carbamidomethylated C) were completely coeluted in both maps obtained by the single 2.1 x 100 mm or 2.1 x 150 mm column as shown in Figure 3, but they were partially (in half-height) separated by the tandem columns. Meanwhile, Figure 3 also shows that N-deamidated HT6* and light chain tryptic peptide LT14 (VDNALQSGNSDESVTEQDSK) achieved baseline resolution by the tandem columns, which was not the case for the single columns, particularly, for the single 2.1 x 100 mm column. Similarly, as shown in Figure 4, the unmodified heavy chain tryptic peptide HT37 (GFYPSDIAVEWESNGQPENNYK, peak 1 in Figure 4) and its deamidated products (peaks 2, 4, 5, and 6, see Figure 4 for details), which were only partially resolved by single columns (particularly for the 2.1 x 100 mm column), were baseline resolved by the tandem columns.

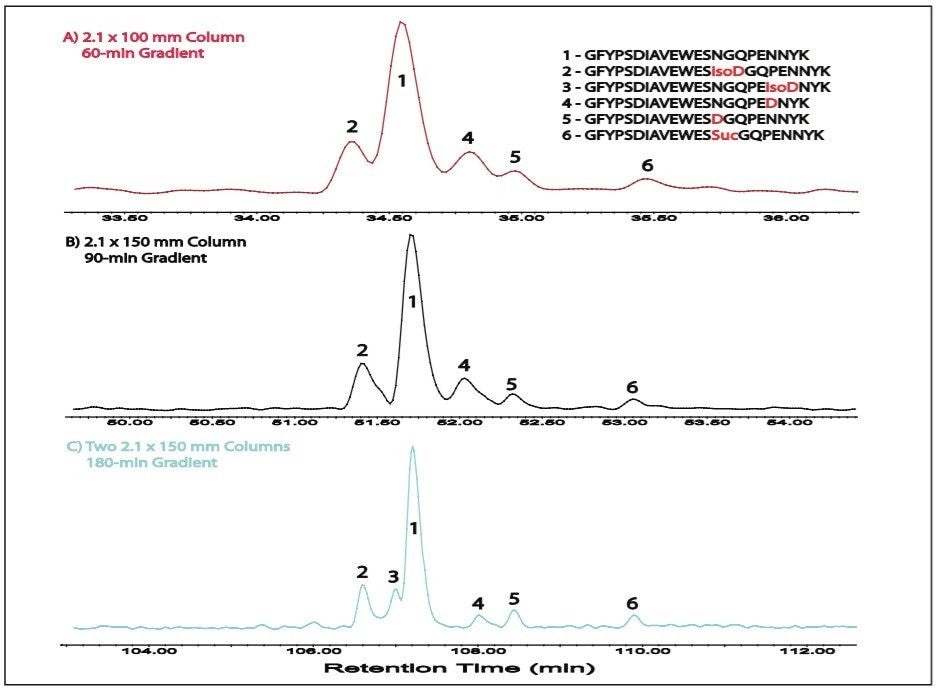
More importantly, the tandem columns separated the deamidated product of peak 3, which was completely coeluted with peak 2 or peak 1 in the maps obtained by single columns. HT37 is also called “PENNY” peptide. It receives particular attention because it tends to be easily deamidated during production and storage, and potentially affects drug quality. The complexity of its deamidation products makes it an analytical challenge. Here, we demonstrated the ACQUITY UPLC System with tandem columns is capable of completely separating modified and unmodified components of “PENNY” peptide.
Mapping of mAb tryptic digests is a typical biopharmaceutical application. By comparing the maps of the mAb tryptic digest obtained at different column lengths, the difficulty of characterizing the structure and modifications of mAbs is shown. Due to the complexity of the resulting digests and the chromatographic challenge of peptide mapping, good chromatographic is paramount. For such applications, the ACQUITY UPLC System configured with a tandem of two 2.1 x 150 columns shows advantages over single (2.1 x 100 mm or 2.1 x 150 mm) column configurations due to the improved resolution and capability to separate unmodified and modified peptides such as N-deamidated peptides.
Meanwhile, using this tandem column configuration, the sequence coverage of the mAb monitored by BiopharmaLynx 1.2 was 97.2% for the light chain and 98.2% for the heavy chain for each of six continuous injections, demonstrating high sequence coverage and excellent reproducibility.
Coelution is a serious challenge for peptide mapping applications. In this work, we demonstrated that the ACQUITY UPLC System configured with a tandem of 2.1 x 150 mm columns is capable of improving resolution of peptide maps and reducing coelution. This improved resolution is particularly useful for identification and quantification of protein heterogeneities with UPLC-UV. This tandem column configuration was implemented on the ACQUITY UPLC System (binary solvent management) with a column temperature ≥ 40 °C and did not require any additional equipments or parts. Furthermore, this instrumental configuration is reproducible and easy to use.
720003362, March 2010