In this application note, we describe the ability of a novel UPLC-MS/MS platform, the Waters Xevo TQ Mass Spectrometer, to monitor potential matrix interferences in plasma while monitoring a pharmaceutical compound of interest.
Development of a fast, sensitive, and robust bioanalytical LC-MS/MS assay is essential for cost-effective and compliant processing of samples in biological fluids.
However, any bioanalytical assay can be hampered by sample matrix effects. Components such as drug metabolites, proteins, and phospholipids within biological matrices frequently interfere with the robustness and sensitivity of the assay. Furthermore, regulatory authorities now require that matrix effects be determined in these assays.
Therefore, it would be very advantageous to actively monitor and characterize the presence of matrix components coeluting with the compound of interest during LC-MS/MS assay method development. Significant time and effort could be saved by ensuring that components in the matrix are well resolved from the analyte of interest. However, conventional tandem quadrupole mass spectrometers cannot acquire multiple reaction monitoring (MRM) data while acquiring full-scan data at speeds fast enough for the narrow chromatographic peaks generated by modern separations techniques such as UPLC.
In this application note, we describe the ability of a novel UPLC-MS/MS platform, the Waters Xevo TQ Mass Spectrometer, to monitor potential matrix interferences in plasma while monitoring a pharmaceutical compound of interest. Unique to the Xevo TQ MS platform is its ability to switch between MS and MS/MS modes in a UPLC run that typically generates peak widths of 2 to 3 seconds.
Alprazolam was spiked into rat plasma at a concentration of 10 ng/mL and then precipitated with acetonitrile using a 2:1 acetonitrile/plasma ratio. The sample was then centrifuged at 13,000 RCF for 5 minutes. The supernatant was removed and injected onto the UPLC-MS/MS system.
|
LC system: |
Waters ACQUITY UPLC System |
|
Column: |
ACQUITY UPLC BEH C18 Column, 2.1 x 50 mm, 1.7 μm |
|
Column temp.: |
40 °C |
|
Flow rate: |
600 μL/min |
|
Mobile phase A: |
0.1 % NH4OH |
|
Mobile phase B: |
MeOH |
|
Gradient: |
5% to 95% B/2 min |
|
MS System: |
Waters Xevo TQ MS |
|
Ionization mode: |
ESI positive |
|
Capillary voltage: |
1000 V |
|
Cone voltage: |
25 V |
|
Desolvation temp.: |
500 °C |
|
Desolvation gas: |
1000 L/Hr |
|
Source temp.: |
150 °C |
|
Scan range: |
m/z 100 to 1000 |
|
Collision energies: |
High 20 V, low 3 V |
|
MRM transition: |
m/z 309 > 281 |

As previously stated, the large number of components found in matrices commonly employed in bioanalysis, such as plasma and urine, can pose a significant problem when developing and validating a quantitative bioanalytical method. Techniques such as solid phase extraction (SPE) and high resolution chromatography are often employed to reduce their effects.1,2
Plasma, for instance, has many endogenous compounds that can interfere with the pharmaceutical compound undergoing quantification. Some of the major interferences in plasma samples that cause ion suppression or enhancement are phospholipids, specifically variants containing the choline head group. Consequently, scientists developing bioanalytical methods often will monitor phospholipids as they can be a major source of matrix effects.
Parent or precursor ion scanning of the indicative choline fragment ion (m/z 184, in positive ion mode) is commonly used to monitor phospholipids (Figure 2).
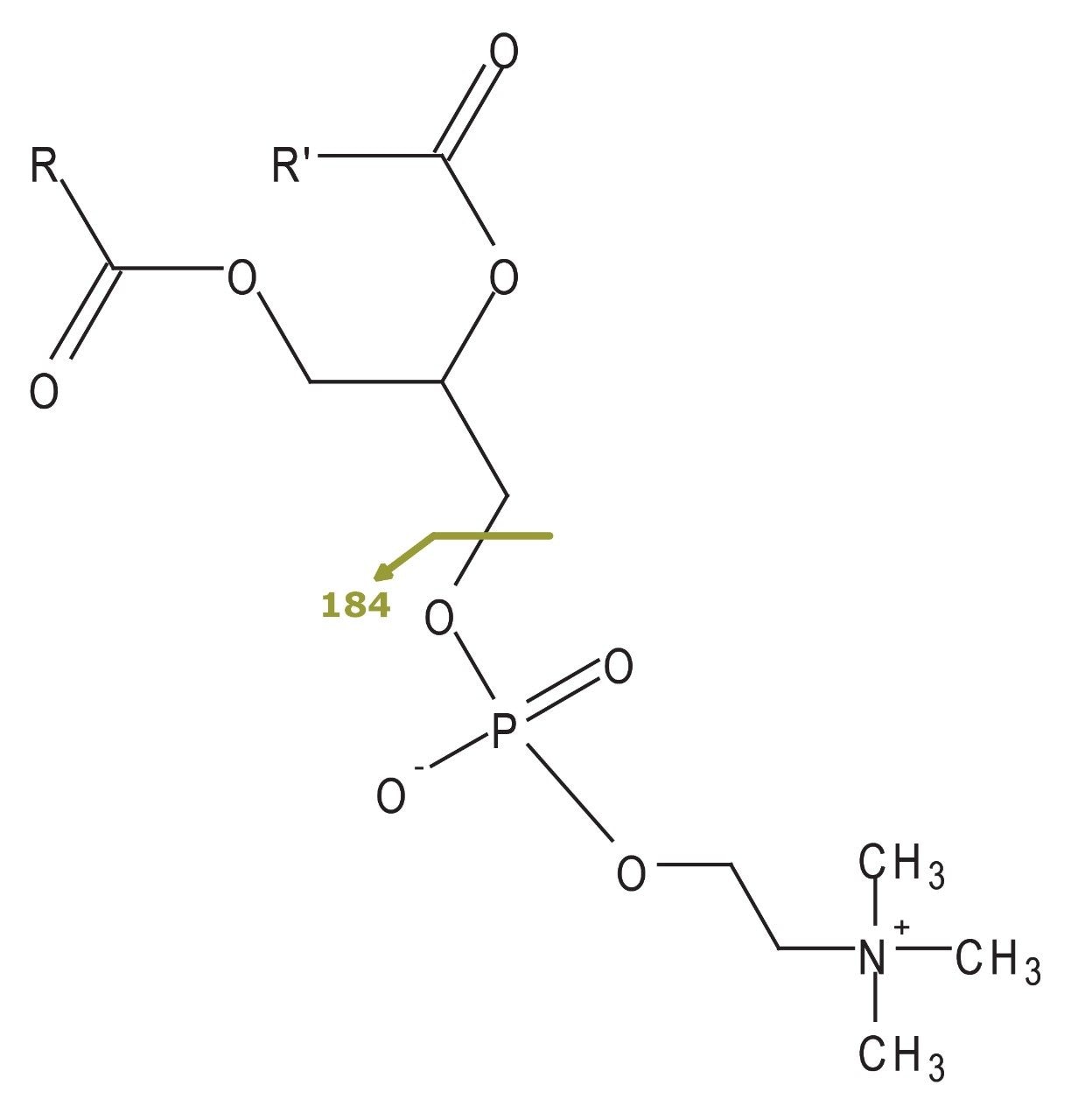
Figure 3 shows the MRM channel for alprazolam above a scan result for precursor m/z 184. In this example, we can see that UPLC methodology facilitates excellent resolution of the analyte of interest from the choline-containing phospholipids.
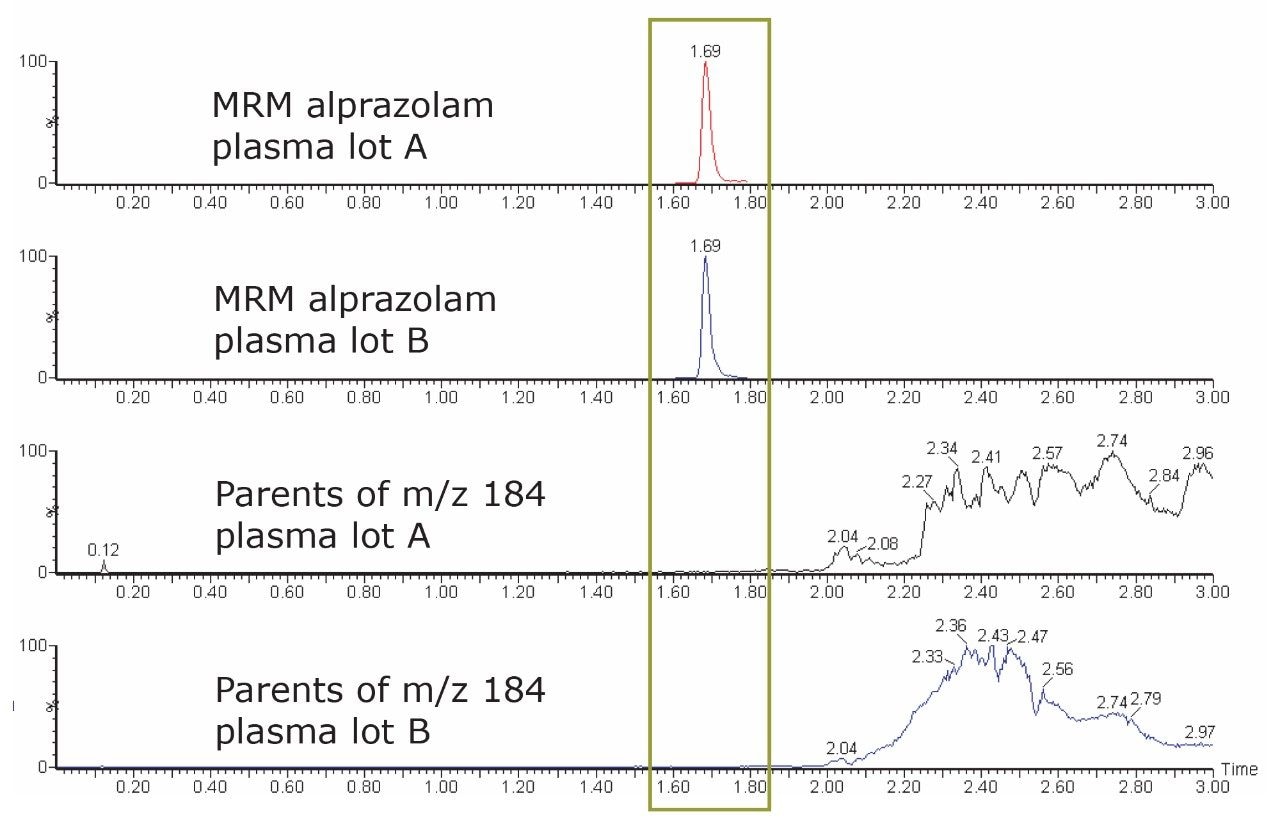
Figure 3 further illustrates the differences between the two scans of the two lots of plasma, indicating that each contains different choline-containing phospholipids. This variance in plasma lot A and lot B exemplifies the reason six different lots of matrix are required to be analyzed during the bioanalytical method validation process.3
Recognizing that there may be other compounds in the matrix that could potentially interfere, and thus become a source of matrix effects, a full MS scan was acquired from the same injection as the MRM channel (Figure 4). Acquiring full-scan data gives the bioanalytical scientist the tools to observe more potential matrix interferences in the samples.
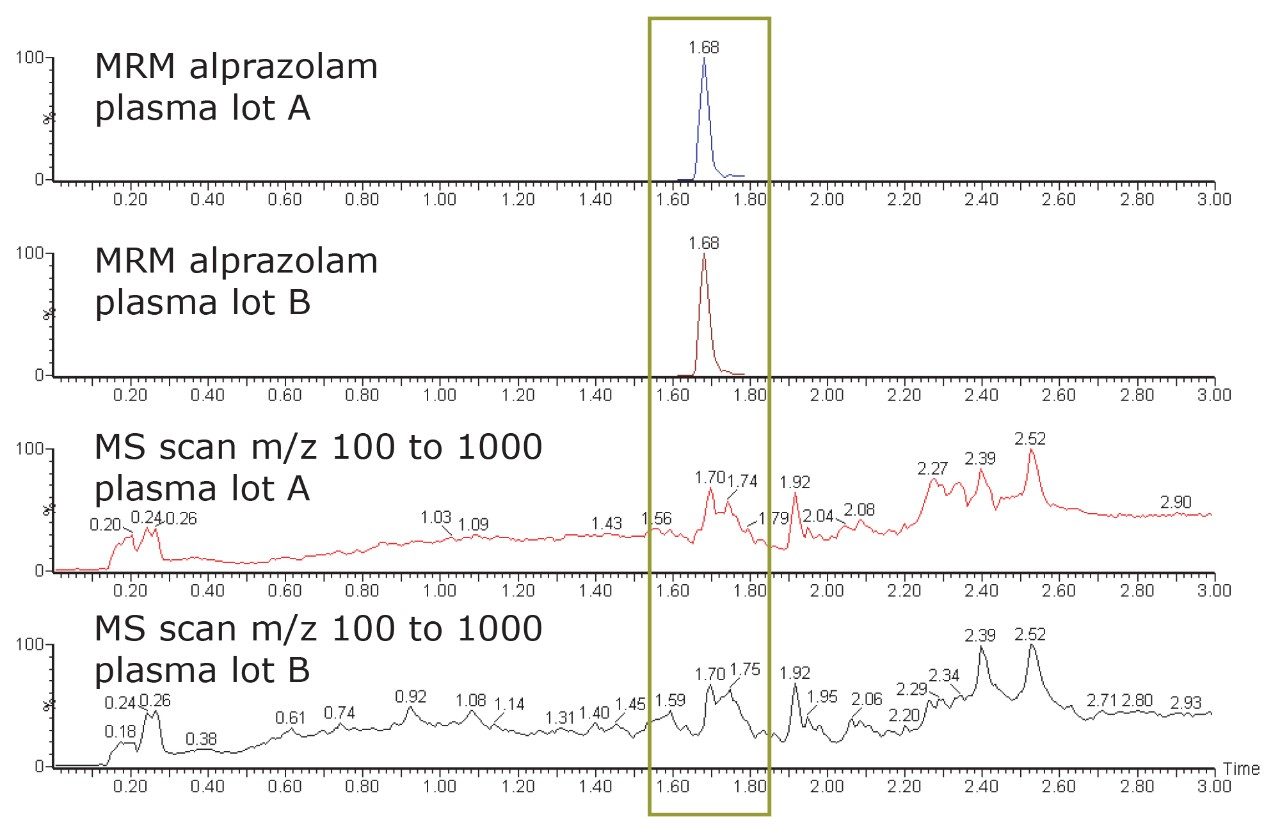
Figure 4 indicates the presence of other coeluting analytes from the matrix. The coeluting analytes have the potential to introduce matrix effects to the analysis of alprazolam, and may impact the quantification of the analyte of interest. The scientist is now in a position to adjust the conditions of the assay method, taking into account the interfering components.
The unique capability of the Xevo TQ MS to rapidly switch between MS and MS/MS modes facilitates simultaneous monitoring of coeluting components as well measuring the compound of interest, even when UPLC peaks are typically 2 to 3 seconds wide.
In this application note, we have shown the novel ability of the Waters Xevo TQ MS coupled with an ACQUITY UPLC System to acquire quality full-scan and MRM data in a single analysis. Using this technique, we monitored potential matrix interferences present in protein-precipitated plasma while monitoring the MRM transition for a model pharmaceutical.
Interferences due to coelution of matrix components can thus be detected “on the fly” in early method development, which reduces:
Thus utilizing the Xevo TQ MS enables researchers to increase the quality of the final MS/MS method while developing a fully-detailed scan record should reviewing the data be of interest.
720002830, October 2008