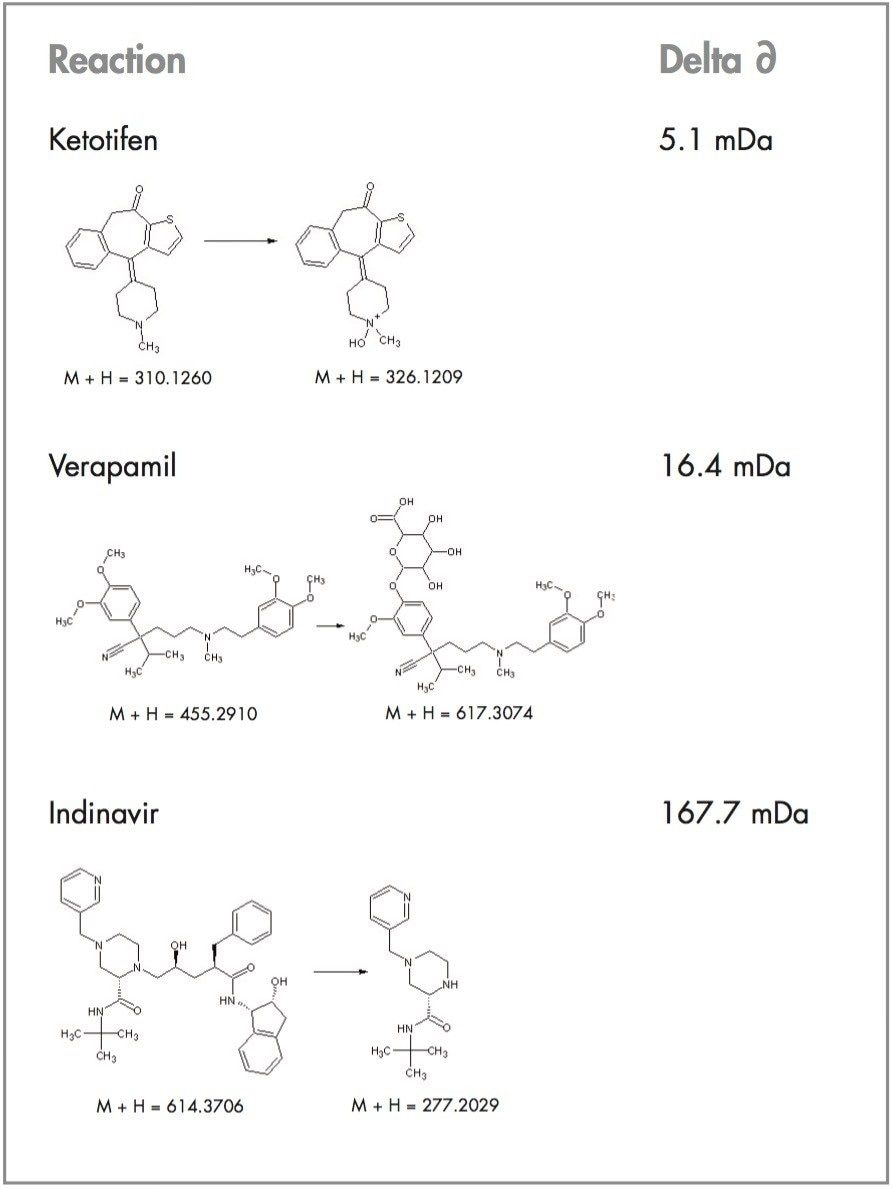
In this study, we concentrate on the in vivo metabolism of verapamil and ketotifen and how we can use the exact mass data filter to remove unwanted false positives.
A rapid and reliable metabolite identification process, even for minor metabolites in complex biological matrices
Within the pharmaceutical and biotechnology industry, LC-MS is widely used as the analytical technique of choice due to its inherent sensitivity and selectivity. The coupling of LC with orthogonal acceleration-time of flight (oa-TOF) mass spectrometers enables scientists to use exact mass information from both MS and MS/MS to confirm and identify metabolites with great confidence. One of the main areas of debate in drug metabolism and major bottlenecks has always been data processing and interpretation of results. In order to address this, a strategy is presented which utilizes the 4 decimal places obtained with time of flight mass spectrometry to mass filter matrix-related peaks.1
An exact mass defect filter2 based on the decimal places of the m/z value of the parent drug under analysis has been developed. It is a post processing filter that removes unexpected metabolite entries in the MetaboLynx Application Manager browser that do not agree with user-defined criteria. The use of this filter can dramatically reduce the number of false entries in the unexpected metabolite table by filtering out the vast majority of matrix-related entries, which may share the same nominal mass as potential metabolites. This allows users to use low threshold values during data processing so that very low metabolite levels are identified without going through the tedious task of manually excluding false positives. It is a very accurate and specific filter because it is based on exact mass and mass deficiencies, which are specific to each parent drug compound of interest.
Each parent drug is comprised of a specific number of elements (C, H, N, O etc.). Depending on the number of each element, the m/z value of the drug of interest will have very specific decimal places. For example, verapamil contains the following elements: C27H38N2O4. This equates to a monoisotopic protonated mass of 455.2910 Da (Figure 1). If we take an alkyl group away (N-dealkylation, a common metabolic route) and add a glucuronide, then the mass is shifted by +162.0164 Da which corresponds to a monoisotopic mass of 617.3074 Da. If we now work out the delta mass difference for the 4 decimal places between verapamil and its N-dealkylated metabolite, we are left with an exact mass deficiency of 0.3074 - 0.2910 = 0.0164 Da. Therefore, if we were to look at a window of around 20 mDa we would be able to detect its N-dealkylated glucuronidated metabolite. Previous knowledge of the metabolites of verapamil is not necessary, as we can make the following assumptions:
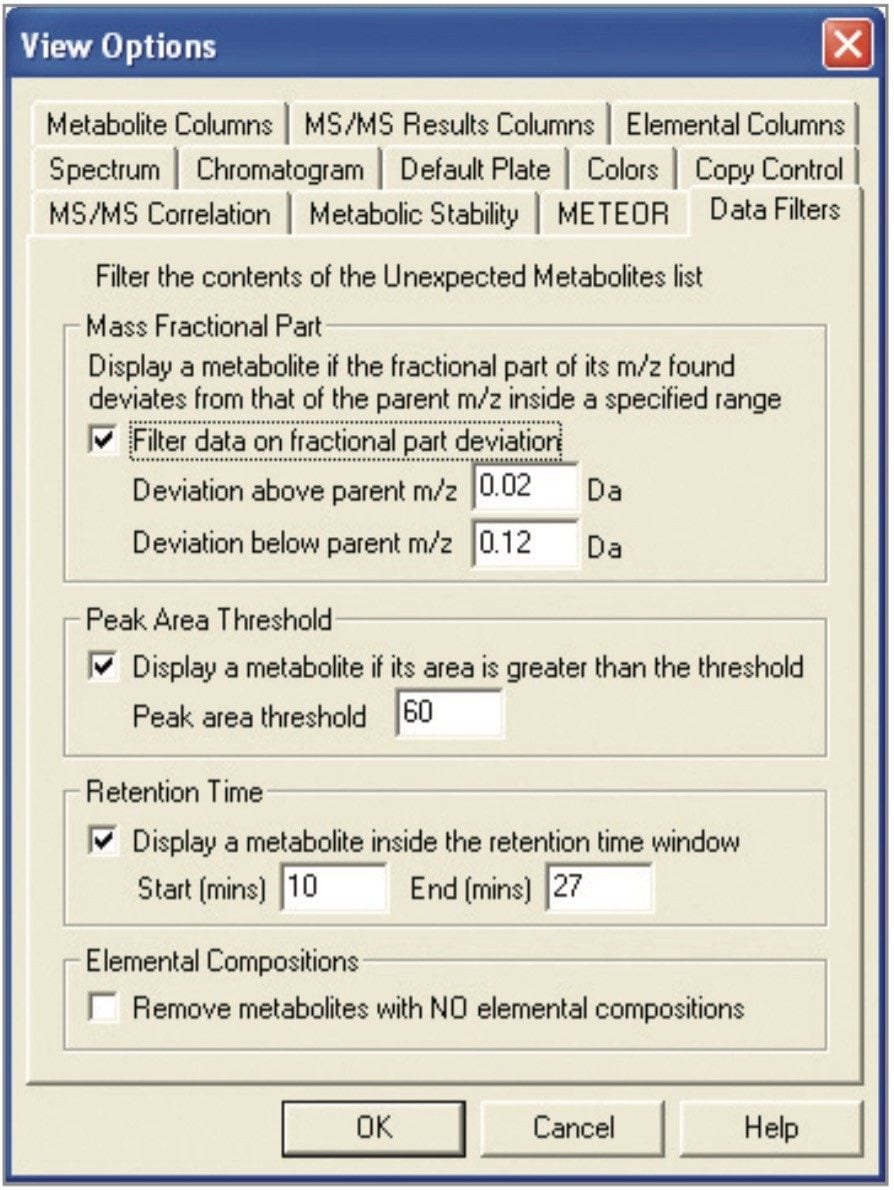
This new development, implemented in the MetaboLynx browser as a data filter, may be applied after the data has been processed (Figure 2). This allows the user to set very low thresholds without the worry of optimizing the set up parameters to detect metabolites because unwanted peaks will be filtered at a later stage.
Furthermore, there are other data filters which can be very useful, such as the peak area threshold and the retention time. With regards to the peak area threshold, the user can set very low area thresholds in order to not miss impor tant low level metabolites. Retention time may also be used to display the portion of the run time where metabolites elute. This option may further remove false positives without reprocessing the data. The retention time window may also be adjusted to display and remove unwanted entries in the unexpected table.
In this study, we concentrate on the in vivo metabolism of verapamil and ketotifen and how we can use the exact mass data filter to remove unwanted false positives.
In vivo metabolism: Lister Hooded rats were administered orally with verapamil and ketotifen (2 rats/compound as a 1mL suspension in 10% acacia) at a dose of 30 mg/kg. Blood was collected (500 μL) 1 hour after administration in Li-heparin tubes. Plasma was prepared by centrifugation and proteins were precipitated with 2 volumes of acetonitrile. The super natant was evaporated, reconstituted in 250 μL of HPLC solvents A/B (70/30, see LC-MS conditions) and injected into the LC-MS system. Urine was collected on dry ice from 0 to 8 h after administration and was injected into the LC-MS system without any dilution. Plasma from rats administered the vehicle only was used as controls. Urine samples from rats administered indinavir were used as control samples for rats administered verapamil.
|
Solvent delivery system: |
Waters 1525μ Binary HPLC Pump |
|
Autosampler: |
CTC Pal |
|
Column: |
Waters XTerra C18, 2.1 x 250 mm, 5 μm |
|
Mobile phase A: |
Ammonium Hydrogenocarbonate 10 mM pH 8.5/Acetonitrile (90/10) |
|
Mobile phase B: |
Ammonium Hydrogenocarbonate 10 mM pH 8.5/Acetonitrile (10/90) |
|
Gradient: |
0–3 min: 0% B 3–40 min: 0–100% B 40–49 min: 100% B 50–60 min: 0% B |
|
Flow rate: |
200 μL/min |
|
Injection volume: |
10 μL |
|
Mass spectrometer: |
Waters Micromass Q-Tof Ultima |
|
Ionization mode: |
Electrospray positive ion mode |
|
Capillary voltage: |
3 kV |
|
Cone voltage: |
35 V |
|
Acquisition range: |
120 870 m/z |
|
Lock mass: |
Verapamil (MH+ = 455.2910) |
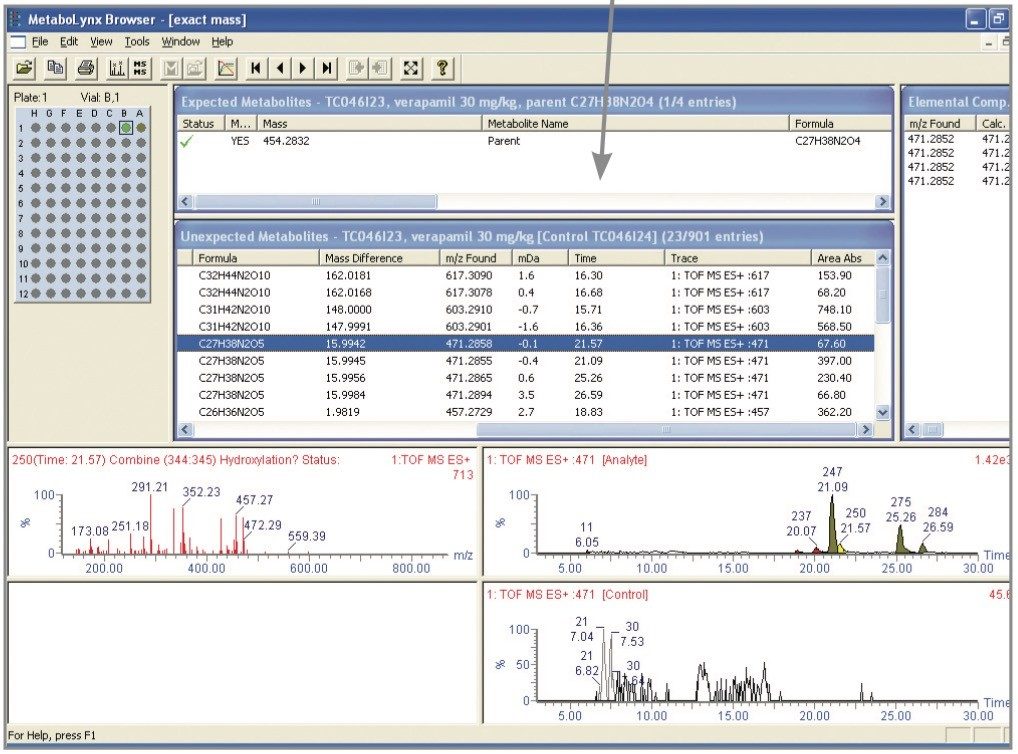
Within the MetaboLynx processing parameters, the integration threshold was set at a peak area of 7 and spectrum intensity at 2%. Only the parent compound was selected as expected. Control and analyte samples were processed using a mass by mass search over the defined mass range (100–700 Da) with a 1 Da step size. The total output from this process was 901 entries identified in the unexpected table of results prior to data filtering with exact mass and peak area threshold. After applying the exact mass data filter (with a 20 mDa mass window above and 120 mDa below the decimals of the parent mass together with an absolute peak area threshold of 60), the number of entries in the unexpected table was reduced to 23 (Figure 3). This dramatically reduced the number of false positives detected, thereby leaving only the most likely potential unexpected metabolites.
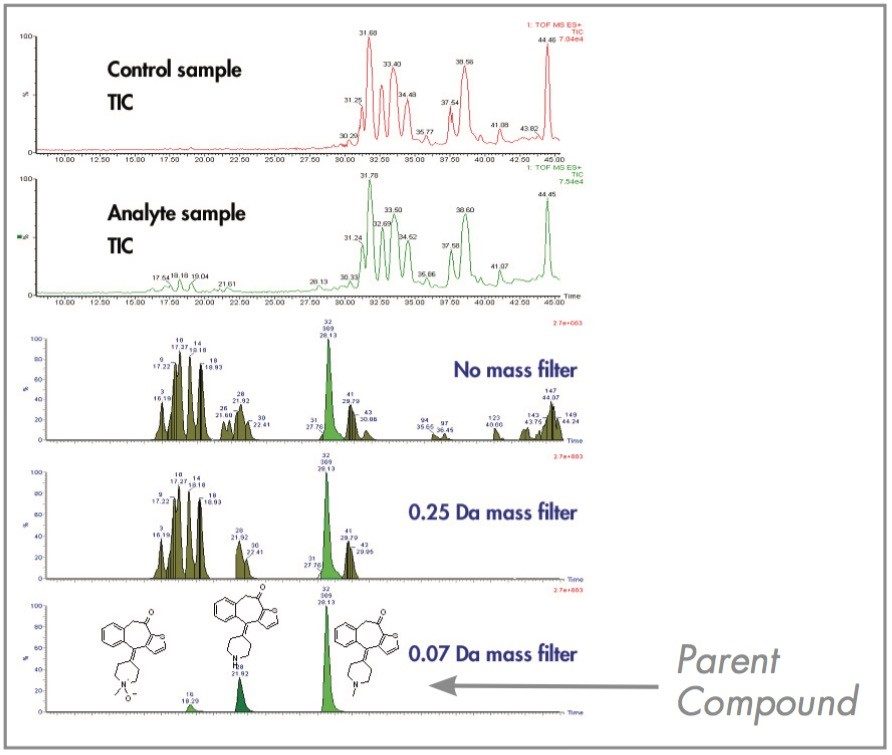
When running plasma samples, data analysis becomes more complicated due to the low ion intensities of circulating metabolites compared to the high ion intensities of endogenous peaks. This is where the exact mass filter can play a very impor tant role in detecting unexpected low level metabolites and removing false positives. As can be seen in Figure 4, when no exact mass filter is applied, a large number of endogenous peaks are detected. This requires time-consuming verification to exclude non-drug related peaks. In the absence of this filter, other experiments such as MS/MS may be required. When the filter is tightened to 250 mDa, fewer entries will remain, and the vast majority of metabolites should fall within this window. Finally, when the window is narrowed to 70 mDa (typical of the single biggest biotransformation possible: glutathione adduct 68.2 mDa), then the two circulating metabolites of ketotifen are very easily detected out of 67 entries. A narrow mass filter was adequate because no cleavage yielding much smaller fragments were expected in this specific case. The advantage of the exact mass filter is that it is a post-processing step, and it can be adjusted to highlight low-level metabolites that fall within criteria set by the user.
Nowadays, rapid turn around of results, especiallyin discover y, is of paramount importance to minimize the “bottleneck” of data processing and reporting. It is impor tant to not only be able to analyze samples and process data quickly, but to also obtain highquality, accurate data from both the analytical and processing steps. These goals are suppor ted with exact mass data acquisition and software algorithms such as the exact mass filter within MetaboLynx. Thefilter allows the user to obtain the best possible data so that decisions on whether the metabolite is real or not is not the rate limiting step. The combined use of mass exclusion from a control sample and subsequent mass filtering using the 4 decimal places from TOF m/z measurements provides a rapid and reliable metabolite identification process, even for minor metabolites in complex biological matrices. “Timing, speed and accuracy of results is a precious asset in today’s fast moving pharmaceutical world.
720001091, March 2005