
The intact mass analysis workflows within the Biopharamceutical Platform Solution with UNIFI enable automated data acquisition, processing, and reporting of a typical method validation sample set. This demonstrates UNIFI Software’s ability to facilitate robust glycoform profiling of a recombinant mAb, removes the necessity of manual data processing, and improves the process of data review and reporting.
The Biopharmaceutical Platform Solution with UNIFI enables a fully integrated workflow for intact mass analysis, including acquisition, processing, and reporting, for organizations in early development and those operating under regulatory compliant environments. The ability to automate and standardize intact mass analysis enables laboratories to deploy their scientific resources with greater efficiency and effect.
Intact mass analysis is a rapid and convenient method for confirming protein identity and profiling product-related variants. In conjunction with other analytical techniques, such as peptide mapping and released glycan analysis, intact mass analysis can help determine if the biomolecule had been correctly cloned, expressed, purified, and formulated during the biopharmaceutical drug development process.
Intact mass analysis can provide a semi-quantitative view of product heterogeneity and is often employed to determine relative composition of product glycoforms. As a lot release test, intact protein mass analysis often provides a quick identity test using the mass of a major variant, sometimes in conjunction with a purity test with defined product variation for peaks corresponding to variants displaying critical product attributes. Demonstration of process consistency through such comparability exercises is critical to obtain initial regulatory approval and for later process improvement studies.
Data processing and report generation often become productivity-limiting tasks for organizations responsible for biotherapeutic protein characterization and analysis. It is still common for LC-MS intact protein data to be manually processed, an inefficient process that lacks standardization and is prone to human error. Further inefficiency and sources of error result from scientists having to reformat results into graphical and tabular formats suitable for communicating information to their organizations.
The ability to automate and standardize data acquisition, processing, and reporting for intact mass analysis allows laboratories to deploy their scientific resources with greater efficiency and effect. The Waters UNIFI Scientific Information System enables these benefits, as well as regulatory compliance, to be realized throughout discovery, development, and quality management organizations.
In this application note, an integrated and compliant-ready solution for intact mass analysis is described. The combination of UPLC separations, optimized application-tested protein column chemistries, the Xevo G2-S QTof for mass detection, all used under control of the UNIFI Scientific Information System, achieves the goal of total workflow automation and standardization.
|
System: |
ACQUITY UPLC H-Class Bio System |
|
Detector: |
ACQUITY UPLC Tunable UV Detector |
|
Column: |
ACQUITY UPLC Protein BEH C4 Column, 300Å, 1.7 μm, 2.1 mm X 50 mm (p/n 186004495) |
|
Column temp.: |
80 °C |
|
Mobile phase A: |
Water |
|
Mobile phase B: |
Acetonitrile |
|
Mobile phase C: |
1% formic acid |
|
Optical detection: |
UV 280 nm |
|
Total run time: |
6.5 min |
|
Time(min) |
Flow(mL/min) |
%A |
%B |
%C |
%D |
Curve |
|---|---|---|---|---|---|---|
|
Initial |
0.40 |
85.0 |
5.0 |
10.0 |
0 |
Initial |
|
1.00 |
0.40 |
85.0 |
5.0 |
10.0 |
0 |
6 |
|
1.01 |
0.20 |
85.0 |
5.0 |
10.0 |
0 |
6 |
|
3.50 |
0.20 |
5.0 |
95.0 |
0.0 |
0 |
6 |
|
3.70 |
0.40 |
5.0 |
95.0 |
0.0 |
0 |
6 |
|
4.00 |
0.40 |
10.0 |
80.0 |
10.0 |
0 |
6 |
|
4.50 |
0.40 |
10.0 |
80.0 |
10.0 |
0 |
6 |
|
5.00 |
0.40 |
85.0 |
5.0 |
10.0 |
0 |
6 |
|
5.50 |
0.40 |
85.5 |
5.0 |
10.0 |
0 |
6 |
|
Mass spectrometer: |
Xevo G2-S QTof |
|
Capillary: |
2.5 kV |
|
Sampling cone: |
80 V |
|
Extraction cone: |
4 V |
|
Source temp.: |
150 °C |
|
Desolvation temp.: |
350 °C |
|
Cone gas flow: |
0 L/h |
|
Desolvation gas flow: |
800 L/h |
UNIFI Scientific Information System
Results derived from an intact IgG1 mAb mass analysis are used to illustrate how this integrated system solution can help the biopharmaceutical laboratories to streamline a common analytical workflow, shown in Figure 1, and more quickly and efficiently communicate key information needed to bring better molecules to market faster.
Waters Intact mAb Mass Check Standard (p/n 186006552) was analyzed by solubilizing the standard (10 mg/mL or 67 μM, 100 μL DI water to standard vial, 5 min sonication), and diluting 20X (Final 3.3 μM, 0.50 μg/μL) with eluent A for Xevo G2 QTof analysis or 200X (0.33 μM, 0.05 μg/μL) for Xevo G2-S analysis.
An automated mAb LC-MS analysis set of 11 injections was automatically acquired, processed, and reported as specified in a single UNIFI method. Data are representative of a simple method development set, where the goal of the researcher is to assess the extent of product glycovariation and determine analytical reproducibility.
For the processed results, a single injection is represented in the review panel of the UNIFI analysis center, shown in Figure 2. This panel is configured to convey chromatographic information (integrated total ion chromatogram), the MaxEnt deconvoluted MS spectrum corresponding to the summed spectra under the detected peak, and a component summary window filtered to display the top five most intensely assigned glycoforms (G0F/G0F, G0F/G1F, G1F/G1F, G1F/G2F, or G2F/G2F).
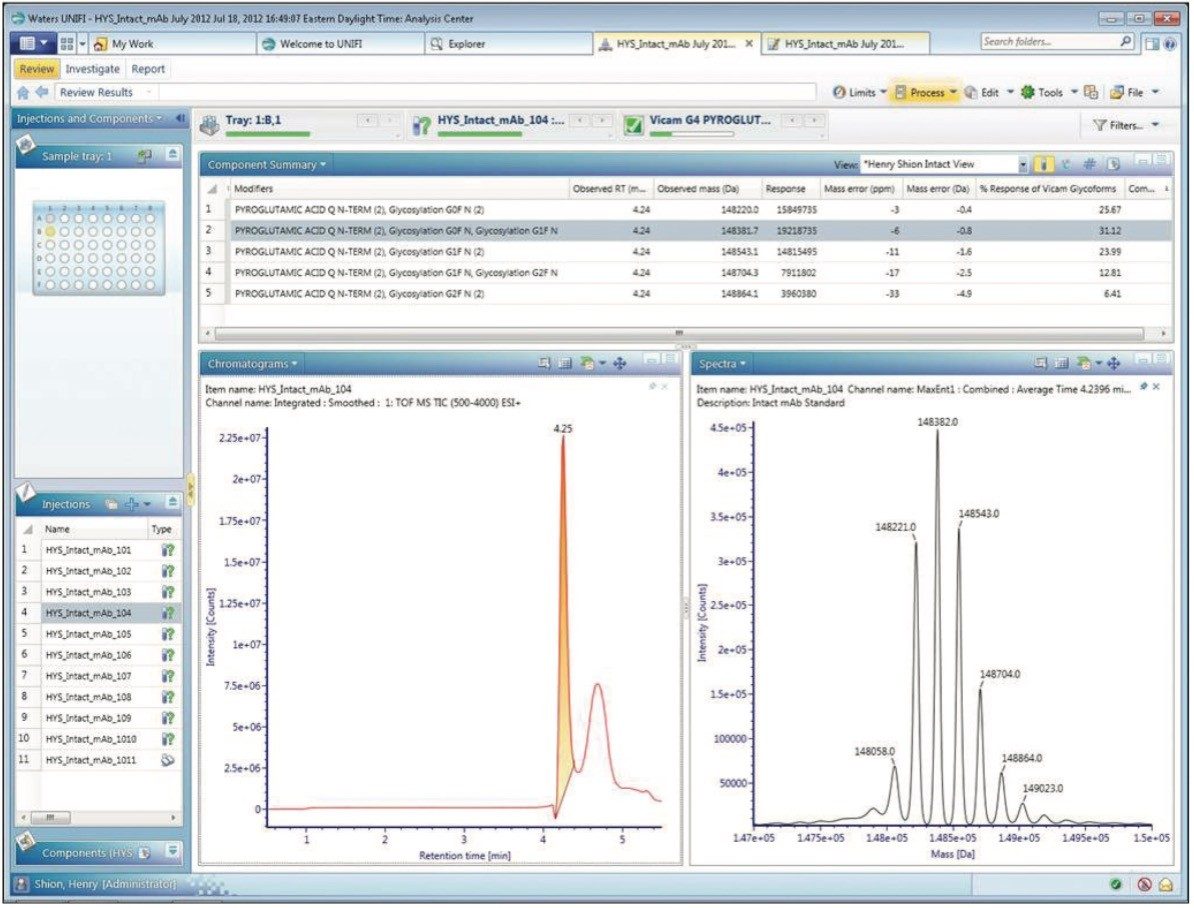
This combined panel enables a researcher to assess chromatographic quality, the quality of MS data processing, and the quality of glycoform assignments in a single display. Closer examination also reveals the relative abundance of each glycoform was automatically calculated as part of the processing.
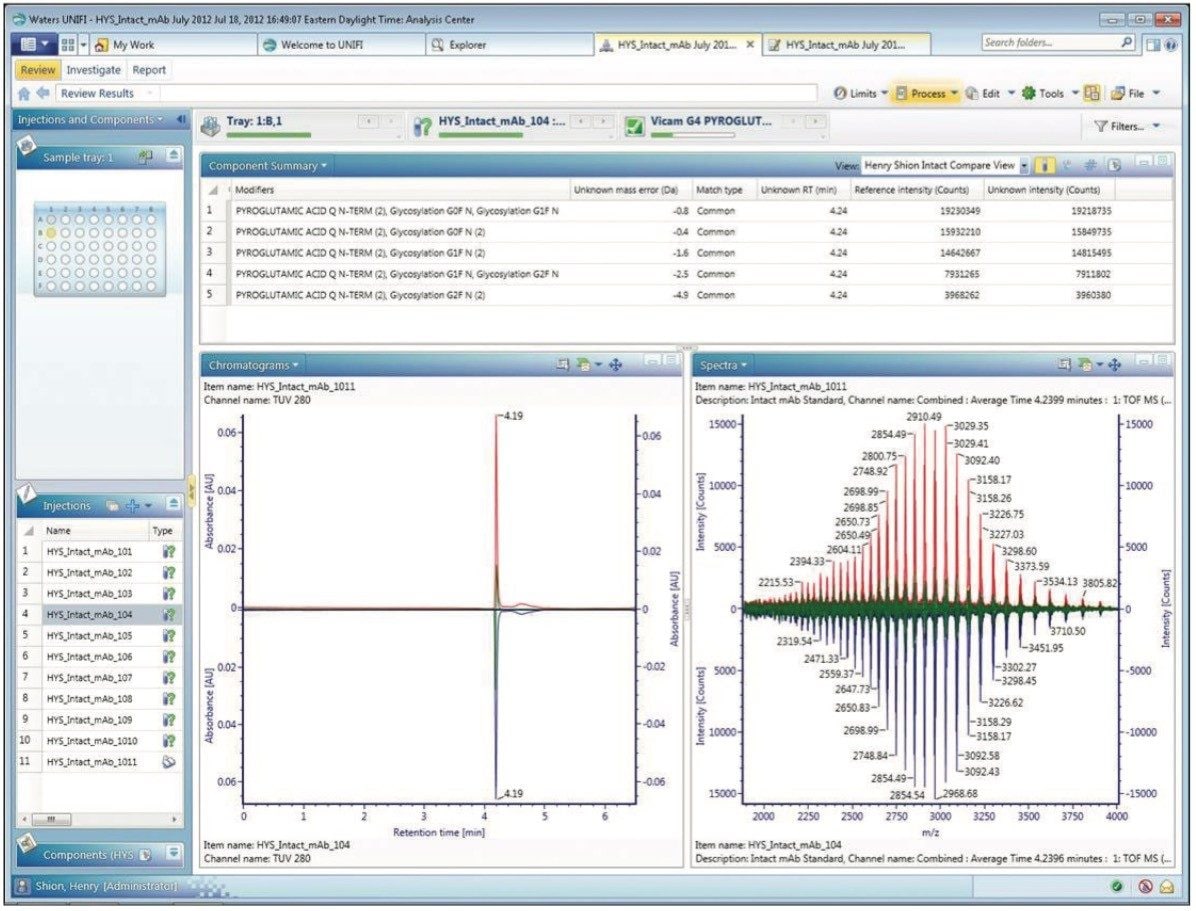
Having designated one sample as the reference enables a researcher to select the comparative mode display of the review tab. The binary comparison display, shown in Figure 3, provides a means to visually examine the differences between the two samples, thus revealing the extent of variation between samples. In this display, comparative chromatograms and spectra (A280 and summed m/z spectra) are depicted, along with the component summary, now reformatted to address comparative questions. Since both injections were from the same sample, minimal experimental result differences are predictably observed.
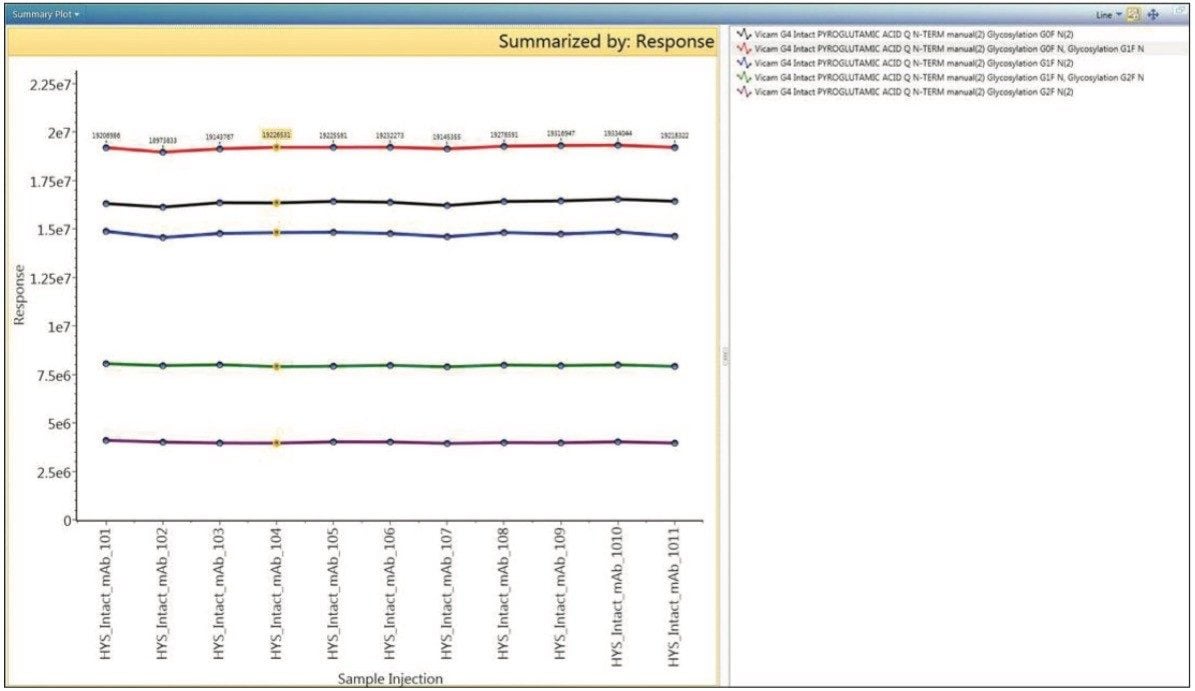
The summary plot tool within the review tab enables researchers to quickly compare trends and differences within the larger data set. The variation of mAb glycoform MS response, as shown in Figure 4, would be a common application of this capability, as would comparisons of observed retention time or mass error across the sample set. The consistent MS response of glycoforms across all injections illustrates the expected reproducibility of the intact mass analysis of replicated injections.
The reporting functionality within UNIFI Software is powerful, addressing one of the common bottlenecks encountered by organizations when generating and managing large volumes of complex scientific data. The ability to customize common report objects by means of filters, formatting, and the use of custom fields and calculations enables report content to be automatically generated by an entire organization with high quality on a consistent basis. Based on the analytical objectives, one or more report templates can be attached to the analysis method.
The first page of a typical intact mass analysis experimental report contains a summary of sample information and acquisition status, as shown in Figure 5. More detailed experimental results (such as TUV and TIC chromatograms, raw and deconvoluted MS spectra, and identified component response summary table) are often grouped for each injection, as shown in Figure 6.
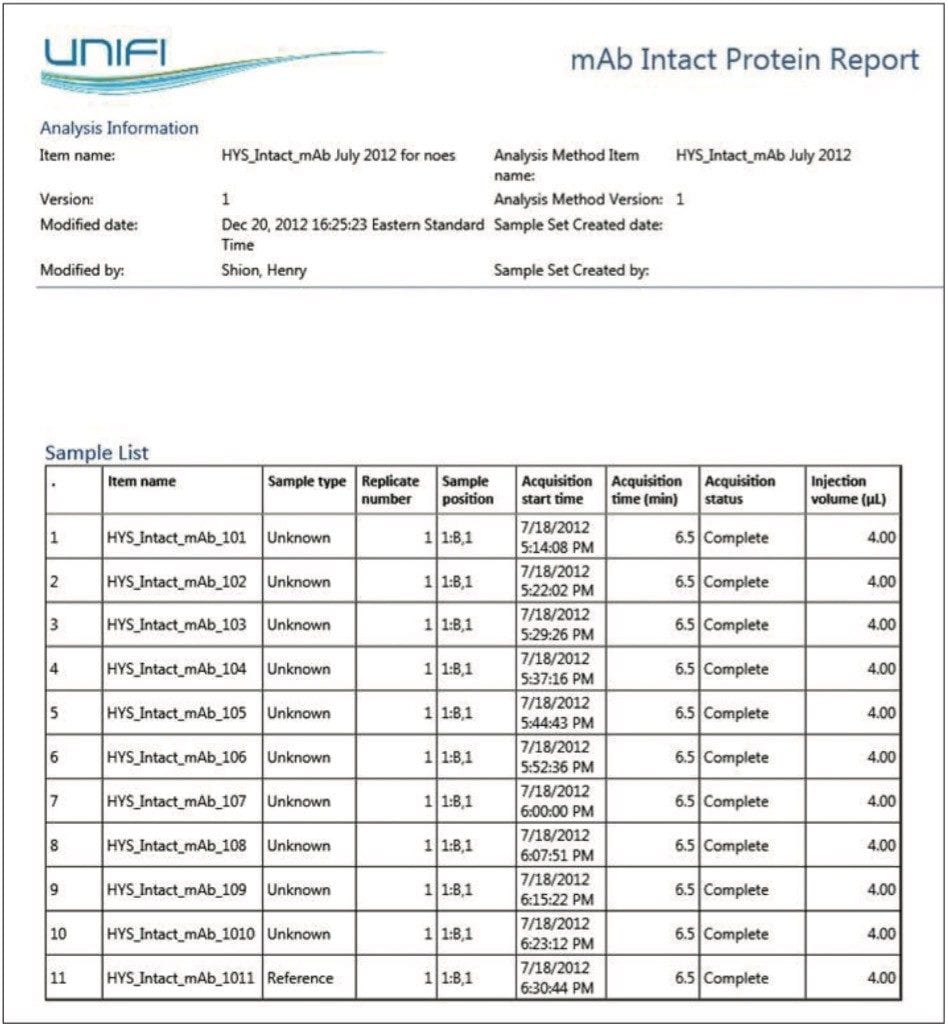
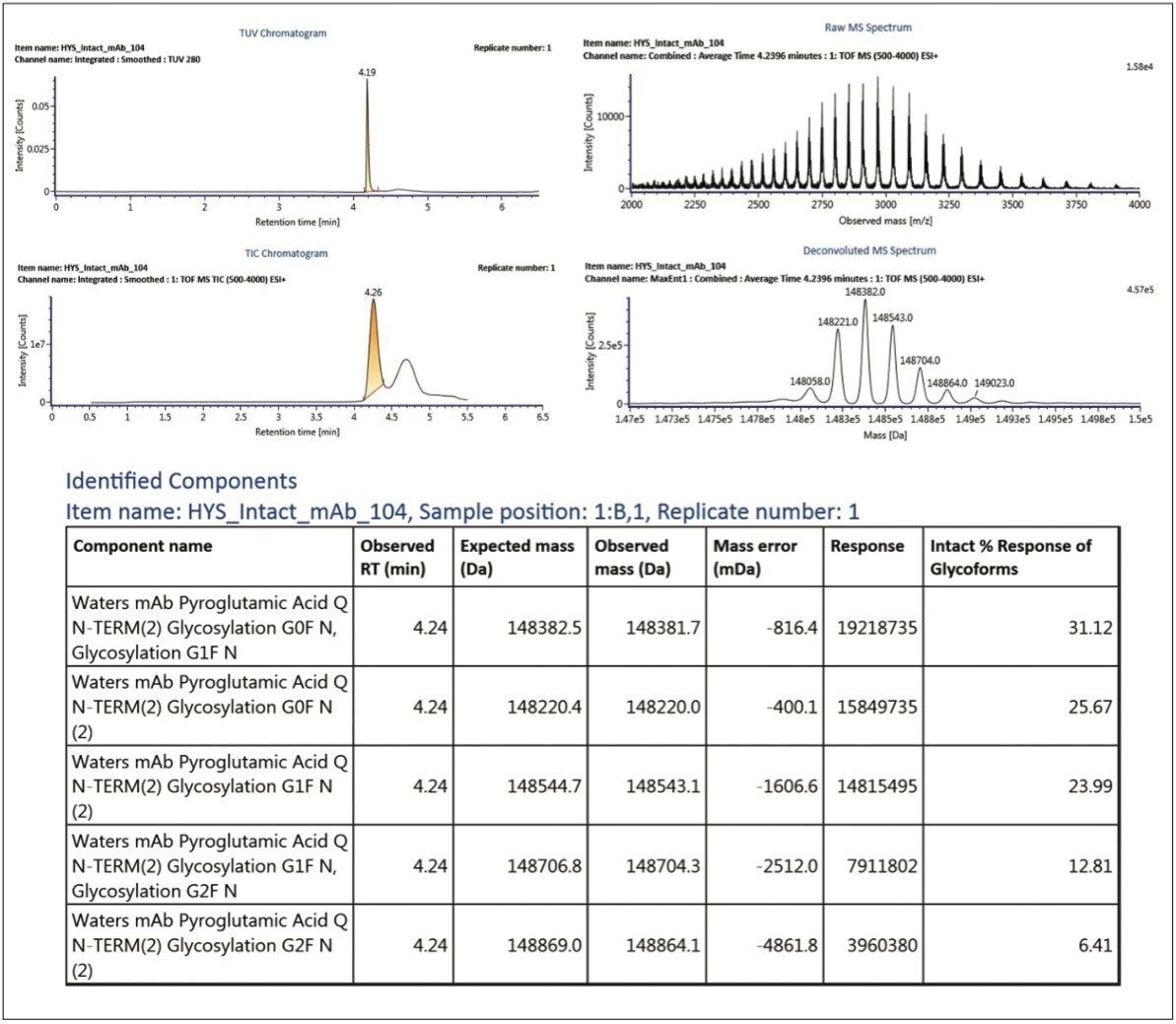
In the case of mAbs, generic report objects were tuned to account for the rapid desalting LC-MS method that was used, the acquisition of UV and MS data, and the typical input m/z and output mass ranges encountered during antibody ESI mass analysis.
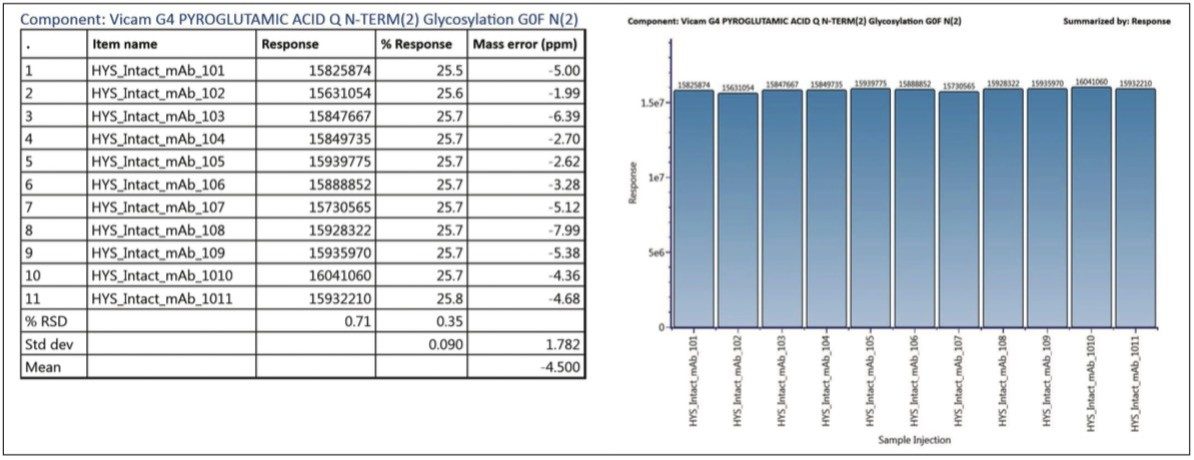
In addition, the ability to automate reporting summary results across the sample sets eliminates the use of external software for data aggregation, as shown in Figure 7. This not only greatly increases the timeliness of communicating results, but avoids the human errors and validation efforts that cost analytical organizations time and money. In the case of this typical method validation injection set, the precision of MS response and mass accuracy is reported for one of the observed glycoforms.
The intact mass analysis workflows within the Biopharamceutical Platform Solution with UNIFI enable automated data acquisition, processing, and reporting of a typical method validation sample set. This demonstrates UNIFI Software’s ability to facilitate robust glycoform profiling of a recombinant mAb, removes the necessity of manual data processing, and improves the process of data review and reporting. The implementation of such highly automated workflows should enable biotherapeutic development and quality organizations to handle larger volumes of sample requests with the same resources, while improving the quality of the information they provide.
720004617, January 2016