For research use only. Not for use in diagnostic procedures.
This is an Application Brief and does not contain a detailed Experimental section.
This application brief illustrates a complete MALDI imaging biomarker discovery workflow through visualization of the effects of various diets on the distribution of Biomarkers of Interest (BOIs) in the rat brain.
Gain a better understanding of physiological processes and pathophysiology of diseases with High Definition MALDI Imaging.
MALDI is one of several ionization methods that enables mass spectral analysis directly from the sample surface of fresh, unfixed tissues that are difficult to access. The use of MALDI to image tissue sections is gaining popularity, which promises to deliver a complete and accurate structural picture of the tissue for putative biomarker characterization and drug development. Not only does this technique allow determination of BOIs, but it also shows their localization with no anatomical distortion. With very little tissue manipulation and disruption, MALDI provides an important advantages for drug development and for the understanding of (metabolic) diseases. Undoubtedly, this technique will become very useful for the demonstration of central nervous system effects of diet, cognition, obesity, gut-brain interactions; metabolic diseases such as diabetes, hypertension, Alzheimer’s; and inflammatory conditions.
Most metabolomics or lipidomics biomarker discovery studies use biofluids (mainly blood and urine) in order to evaluate the biological process that occurs in tissues and organs that are not easily accessible. In this technology brief, we demonstrate how MALDI imaging allows direct analysis of BOIs in tissues; the workflow is shown in Figure 1. This novel approach could lead to a better understanding of the physiological processes and the pathophysiology of diseases because it allows both discovery and localization of biomarkers.
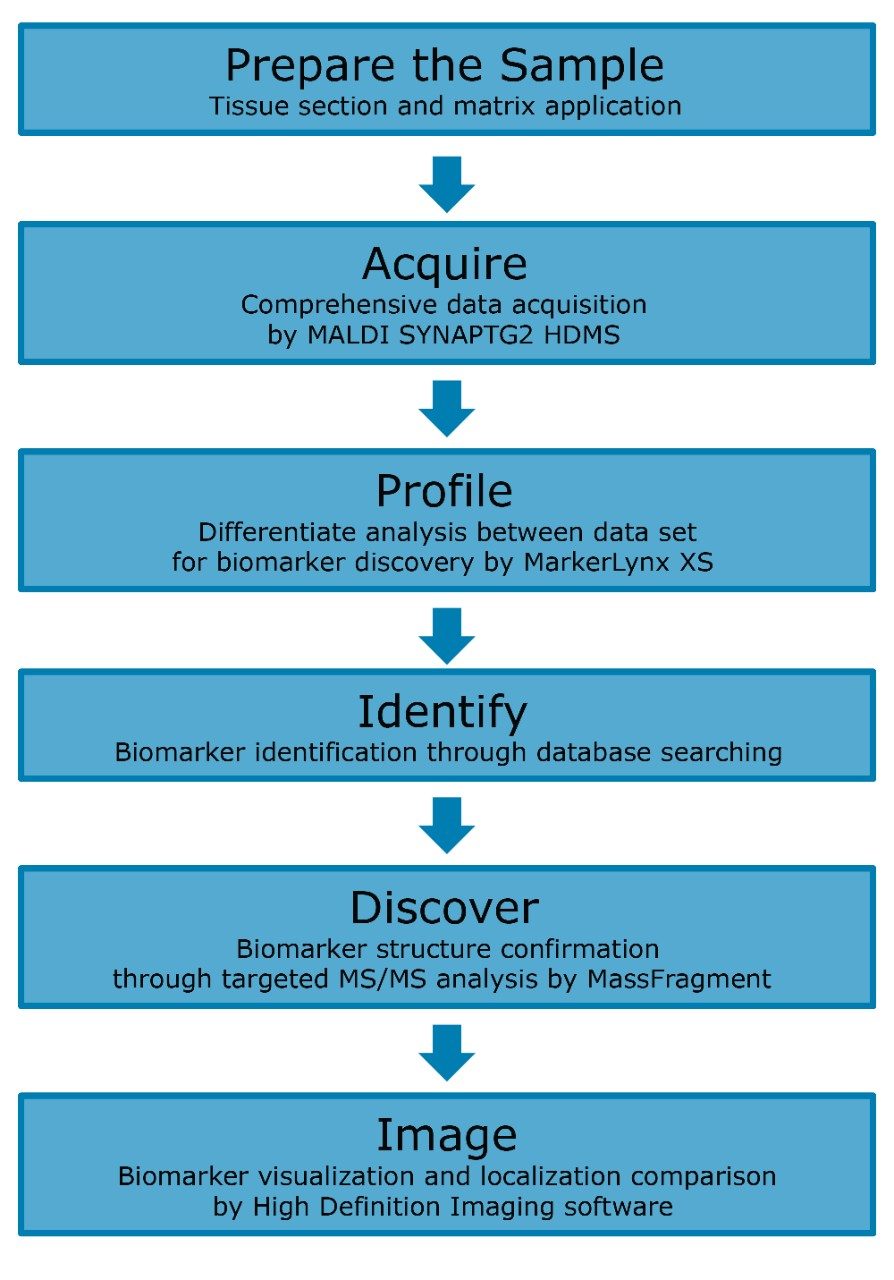
The left brain hemispheres of two rats kept on different diets were sliced in 10 μm sections using a cryostat. Tissue sections were mounted on microscope slides, frozen, and stored at -80 °C until use. 2,5 dihydroxybenzoic acid (DHB) was used as the matrix and a TM Sprayer was used for the matrix application. MALDI imaging analysis was performed on the Waters MALDI SYNAPT G2 HDMS. Positive HDMS full scan data were acquired for the mass range of m/z 100 to 1000. The Nd:YAG laser was operated at a firing rate of 200 Hz with a spatial resolution of 75 μm. Waters new High Definition Imaging Software (HDI) was used for the visualization of MALDI imaging data.
After data acquisition, the complex data set was processed by HDI software and analyzed using MarkerLynx XS Software. The discovered BOIs were identified by database searching using exact mass molecular ion. The identity of the BOIs was confirmed and validated by performing targeted MS/MS experiments, combined with MassFragment Software to propose assignments for the precursor and fragmentation peaks, and matching them between the data generated from the standards and directly from the tissue sample.
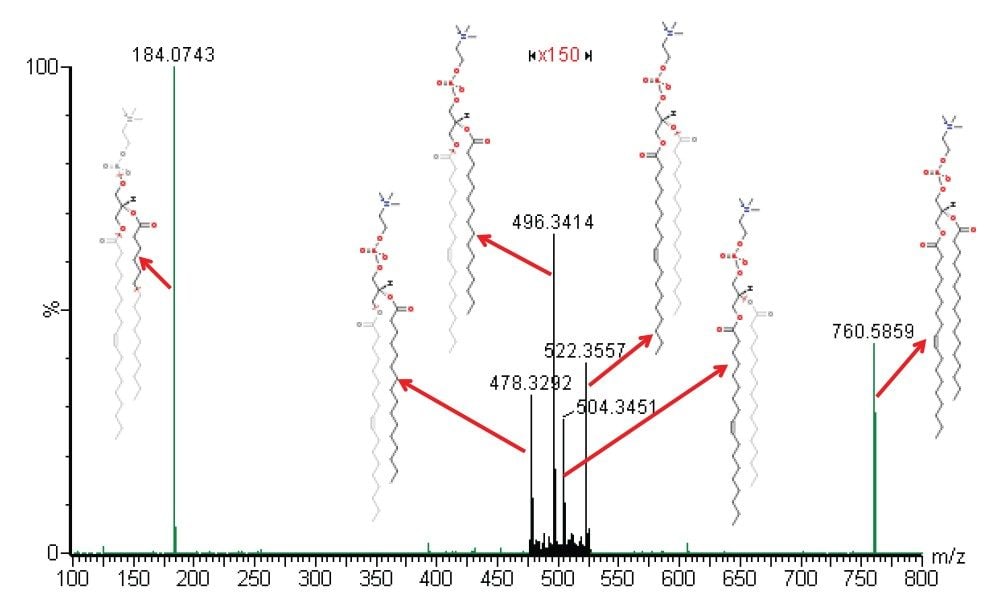
Visualization was performed and compared in HDI using both exact mass and drift time information. Figure 2 shows the MS/MS spectrum of an indentified BIOs, a lipid phosphatidylcholine (PC) (18:1 (9Z)/16:0) or PC (16:0/18:1 (9Z)) observed at m/z=760.5859. The structure of the lipid was confirmed by matching the diagnostic fragmentation pattern of the spectrum with the standard one. The average mass error for the six diagnostic ions in the MS/MS spectrum was 0.48 mDa. Figure 3 shows the MALDI image comparison for the same lipid from the brain tissues of the two rats fed by different diets. Very different distributions of the lipid are observed in the two images, indicating that the found lipid biomarker may be of importance.1 Using this technology, it was possible to highlight several BOIs reflecting the impact of the different diets.
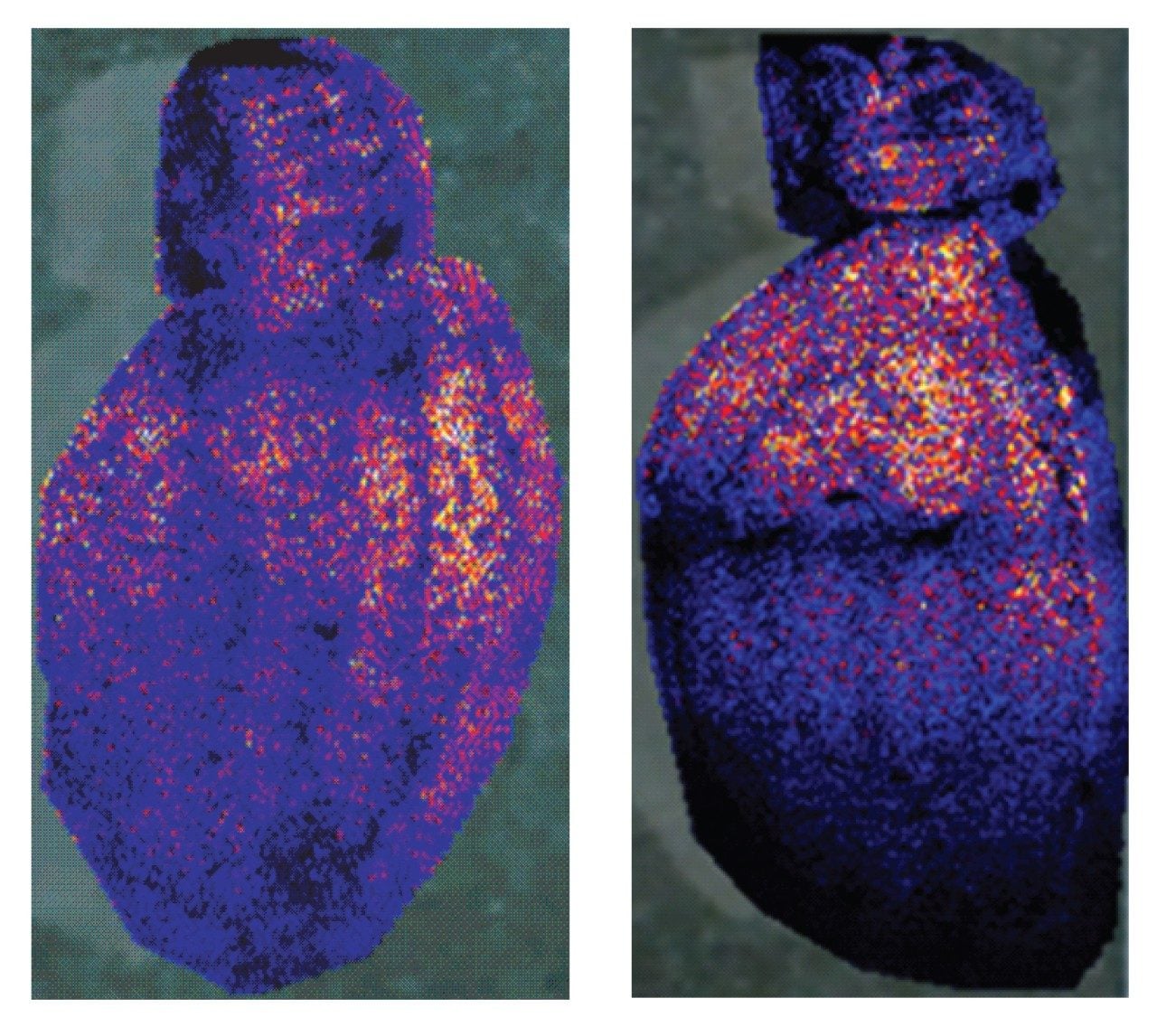
This application brief demonstrates a complete MALDI imaging biomarker discovery workflow for use in rat brain tissues and its modification by dietary components.
Waters High Definition Imaging (HDI) Software integrates pattern generation, HDMS data processing, and visualization in a single interface.
In high definition MALDI imaging, the combination of MarkerLynx XS and MassFragment software enable discovery, identification, and confirmation of BOIs within tissue samples.
Compared to traditional biomarker discovery analysis (mainly LC-ESI-MS approach), this workflow has the potential to lead to a better understanding of physiological processes, as well as the pathophysiology of diseases, with added information about their spatial distribution in heterogenous, not easily accessible tissue sample.
This approach may be applicable to biomarker discovery and distribution analysis in areas including metabolomics, lipidomics, and proteomics in various application fields such as: metabolic diseases, aging, gut-brain interactions, cognition, and inflammatory diseases associated to the health benefits linked to adequate nutritional interventions.
720004135, October 2011