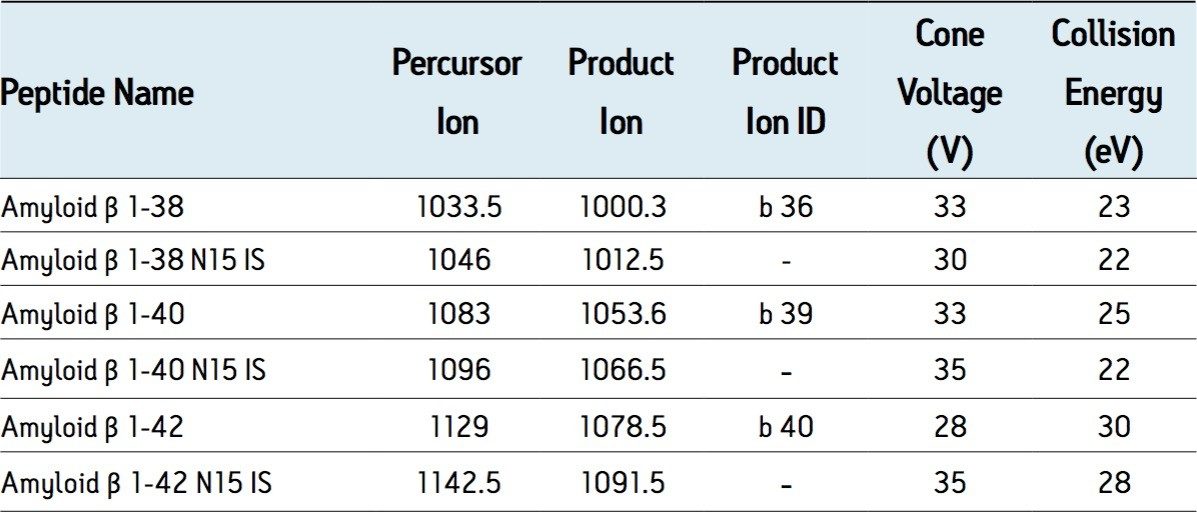
This work focuses on the development of UPLC, MS, and selective SPE sample preparation methods for the 1-38, 1-40, and 1-42 fragments of APP, in support of preclinical studies.
Fast, flexible platforms for peptide quantification are needed, particularly for a discovery setting. This type of methodology would be especially advantageous in the case of amyloid beta (aβ) peptides. The deposition/formation of insoluble aggregates, or plaques, of aβ peptides in the brain is considered to be a critical event in the progression of Alzheimer’s Disease (AD) and thus has the attention of many researchers.
Historically, quantification of aβ peptides in biological fluids has relied mainly on the use of immunoassays, such as ELISA. These assays are time consuming and expensive to develop, labor intensive, are subject to cross reactivity and an individual assay is required for each peptide. In order to meet the throughput requirements and constant flow of demands for new peptide methods in a discovery setting, there is a need for a highly specific yet flexible methodology based on an LC-MS/MS platform. In this work, this platform is coupled with selective sample preparation for the simultaneous quantitation of multiple aβ peptides. This work focuses on methods for the 1-38, 1-40, and 1-42 aβ peptides, in support of preclinical studies.
Development of a bioanalytical method for these peptides is further complicated by their propensity for aggregation, formation of oligomers, poor solubility, nonspecific binding, and hydrophobicity.
As aβ peptides may be present at very low concentrations, we developed a solid-phase extraction (SPE) sample preparation protocol to enrich the amyloid beta fraction in CSF. The SPE method concentrates the sample to improve detection limits while eliminating matrix interferences and optimizing solubility of the aβ peptides in the mass spectrometer injection solution. A high throughput, high resolution UPLC-MS/MS quantitation method was also developed.
This work focuses on the development of UPLC, MS, and selective SPE sample preparation methods for the 1-38, 1-40, and 1-42 fragments of APP, in support of preclinical studies. Sequence, pI and molecular weight (MW) information for these peptides is shown in Figure 1. The use of a single, high throughput assay for multiple aβ peptides- without time consuming immunoprecipitation steps was developed and validated. The speed, selectivity, and specificity of this technique for simultaneously quantitating multiple aβ peptides in CSF are demonstrated.

As strategies emerge for disease modification in AD, the quantification of multiple aβ species (in addition to aβ 38, 40 and 42) that may be linked to AD pathology may help to offer more insight into this disease and its progression. The method described herein shows promise for adaptation to quantitate those peptides as well.
|
Column: |
ACQUITY UPLC BEH C18 300Å, 2.1 x 150 mm, 1.7 μm Peptide Separation Technology |
|
Column Temp.: |
50 °C |
|
Sample Temp.: |
15 °C |
|
Injection Volume: |
10 μL |
|
Injection Mode: |
Partial Loop |
|
Flow Rate: |
0.2 mL/min. |
|
Mobile Phase A: |
0.3% NH4OH in H2O |
|
Mobile Phase B: |
90/10 ACN/mobile phase A |
|
Strong Needle Wash: |
60:40 ACN:IPA + 10% conc. NH4OH (600 μL) |
|
Weak Needle Wash: |
90:10 0.3% NH4OH in H2O:ACN (400 μL) |
|
Time |
Profile |
Curve |
|
|
(min) |
%A |
%B |
|
|
0.0 |
90 |
10 |
6 |
|
1.0 |
90 |
10 |
6 |
|
6.5 |
55 |
45 |
6 |
|
6.7 |
55 |
45 |
6 |
|
7.0 |
90 |
10 |
6 |
|
Capillary Voltage: |
2.5 V |
|
Desolvation Temp.: |
450 °C |
|
Cone Gas Flow: |
Not used |
|
Desolvation Gas Flow: |
800 L/Hr |
|
Collision Cell Pressure: |
2.6 x 10(-3) mbar |
|
MRM transition monitored, ESI+ : |
See Table 1 |
Sample Pre-treatment
200 μL human CSF, monkey CSF, or spiked artificial CSF + 5% rat plasma was diluted 1:1 with 5 M guanidine HCL and shaken at room temperature for 45 minutes. This was then diluted further with 200 μL 4% H3PO4 in H2O.
Note: for spiked samples, samples were allowed to equilibrate at room temperature for 30 min after spiking and prior to dilution with guanidine HCl.
Samples were extracted according to the protocol in Figure 2. All solutions are made up by volume. All steps applied to wells of μElution plate containing samples.

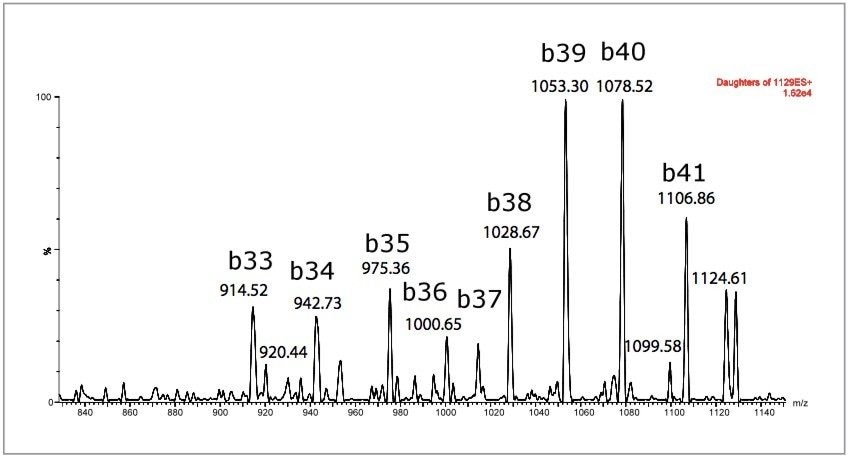
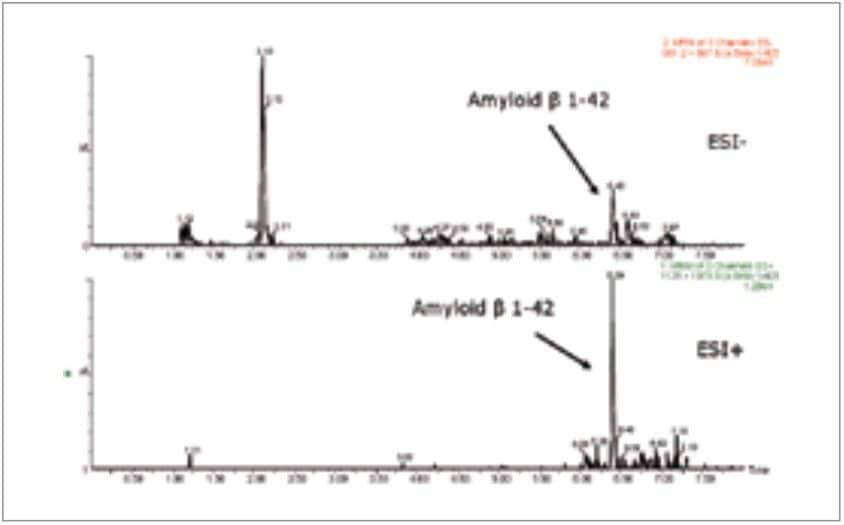
MS was performed in positive ion mode since CID of the 4+ precursor ion yielded several distinct product ions corresponding to specific b sequence ions (representative spectrum shown in Figure 3.) MS/MS in negative ion mode yielded a dominant water loss. Figure 4 demonstrates one example of the specificity difference between both methods in CSF. Although overall sensitivity was higher in solvent standards using the negative ion method, the sensitivity difference was mitigated in the presence of matrix. The improved specificity and signal-to-noise in positive ion mode proved critical for accurate quantitation in CSF samples.
Mass range of the instrument was also an important factor in obtaining specificity. The Xevo TQ MS has a mass range of 2048 on both quads, easily allowing us to choose a more specific 4+ rather than 5+ precursor and fragment pair.
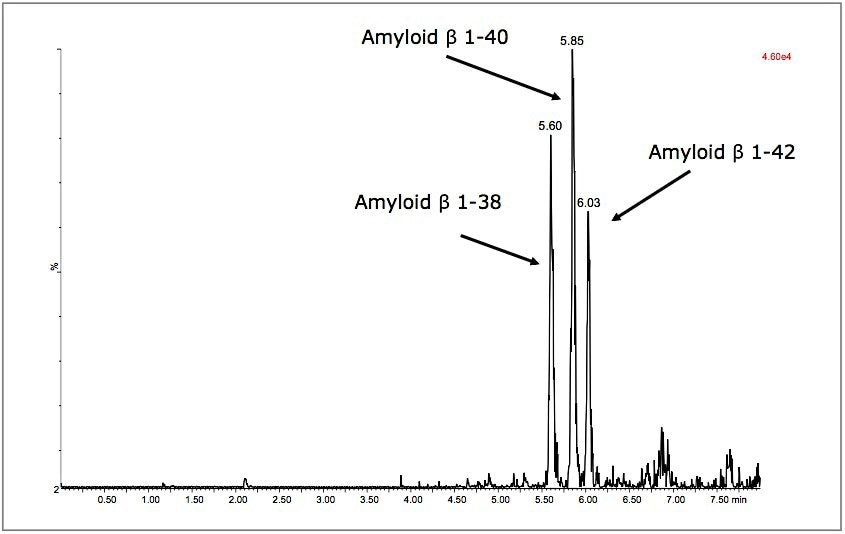
Separation of the three amyloid β peptides is shown in Figure 5. While the exact amount of NH4OH in the mobile phase was critical for negative ion sensitivity, the signal in ESI+ proved to be more robust to subtle changes in mobile phase composition, providing a minimum of >24 hour LC/autosampler stability. In contrast, 50% or more of the ESI– signal was lost after 10-12 hours due to the natural change in NH4OH concentration (volatility) in the mobile phase. This further reinforced the robustness of an ESI+ MS method.
SPE was performed using Oasis MCX, a mixed-mode sorbent, to enhance selectivity of the extraction. The sorbent relies on both reversed-phase and ion-exchange retention mechanisms to selectively separate the aβ fraction from other high abundance polypeptides in complex CSF samples. The Oasis μElution plate (96-well format) provided significant concentration while eliminating evaporation and reconstitution, thus minimizing peptide losses. In addition, binding of the peptides by ion-exchange imparted orthongonality into the overall method as the UPLC separation is performed in the reversed-phase dimension.
During initial method development, a high degree of non-specific binding (NSB) was observed when artificial CSF was extracted. Thus, 5% rat plasma (having a different amyloid β sequence) was added to eliminate the NSB.
The SPE method was one of the more critical aspects of the overall methodology. Very selective isolation of the amyloid fraction coupled with the resolution of analytical-scale flow UPLC, facilitates rapid analysis of pre-clinical samples.
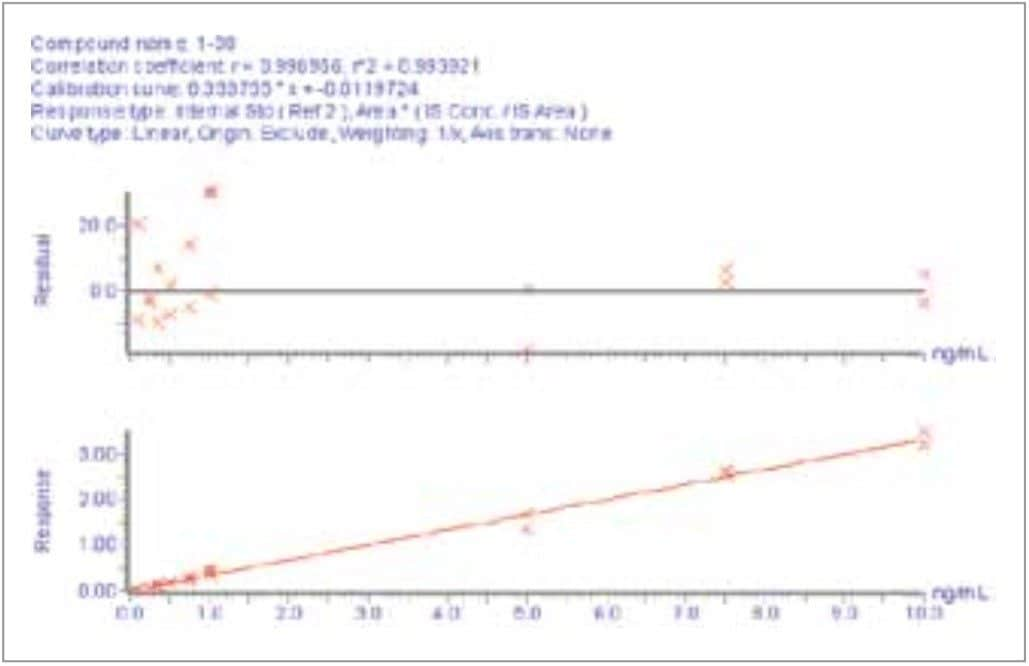
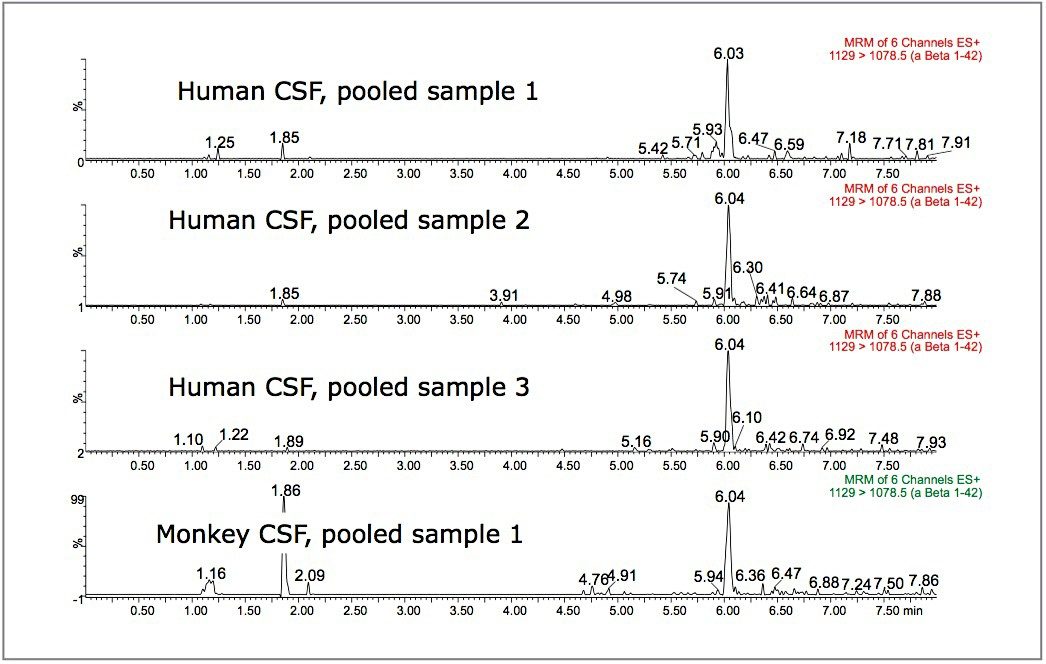
N15-labeled internal standards were used for each peptide. Standard curves for all 3 aβ peptides were linear (1/x weighting) from 0.1 to 10 ng/ mL in artificial CSF + 5% rat plasma. A representative standard curve for amyloid β 1-38 is shown in Figure 6. Basal levels of the aβ peptides were quantitated using both standard curves prepared from over-spiked human CSF and from artificial CSF + 5% rat plasma. Calculated basal levels were not statistically different. The artificial CSF was chosen as it is a less expensive and more readily available matrix. Basal levels of amyloid β 1-42 extracted from 3 sources of human and 1 source of monkey CSF are shown in Figure 7. Statistics for the determination of basal levels of all 3 aβ peptides are shown in Table 2.
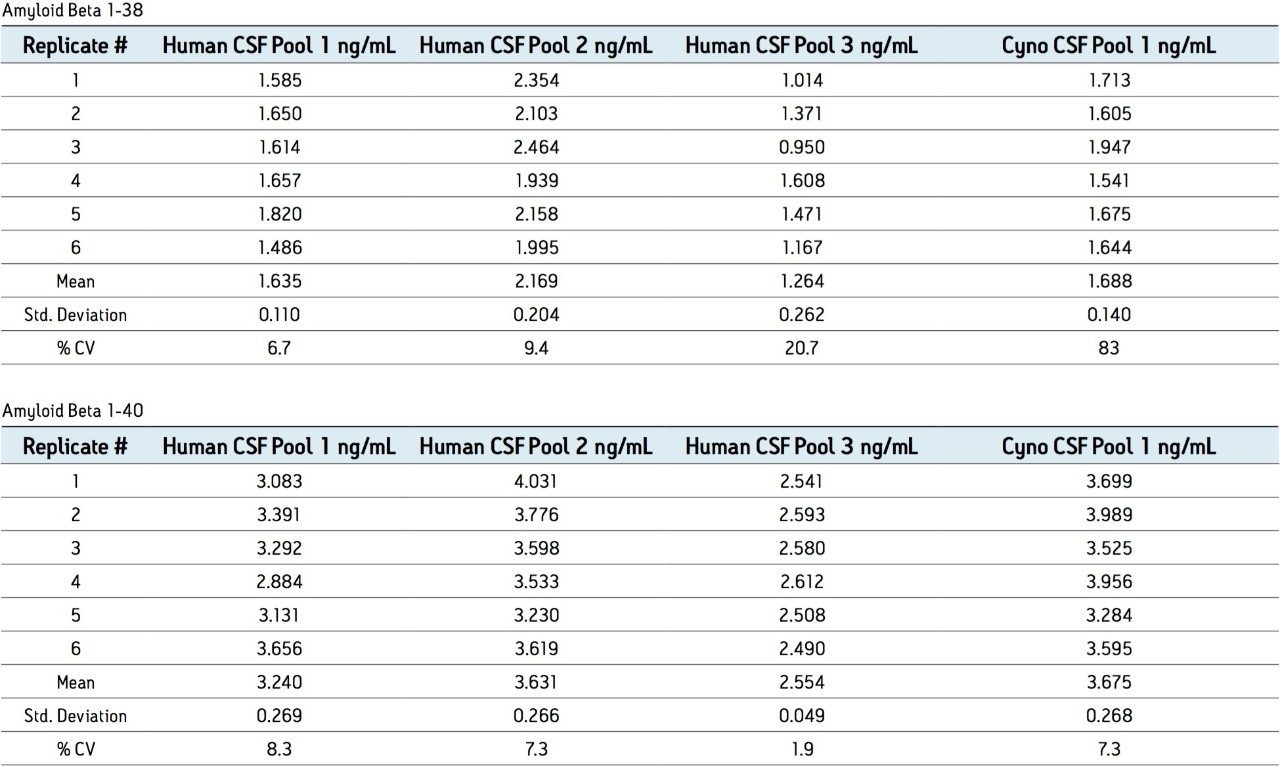
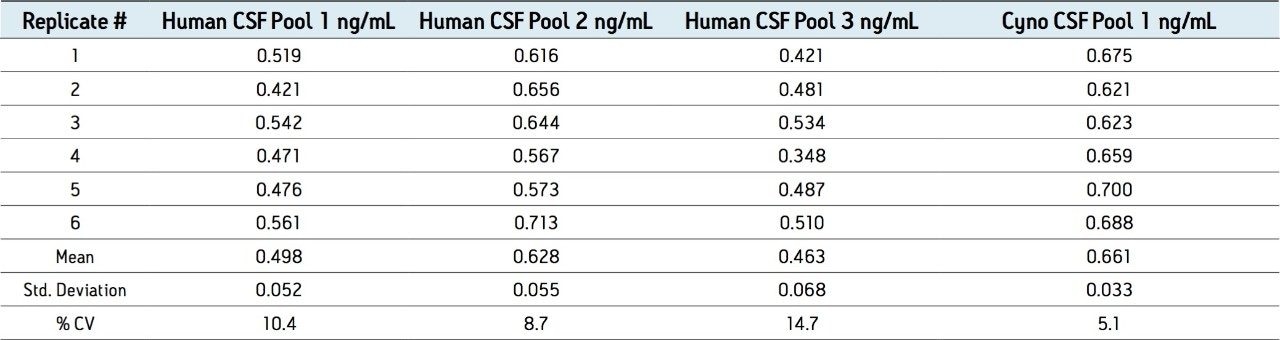
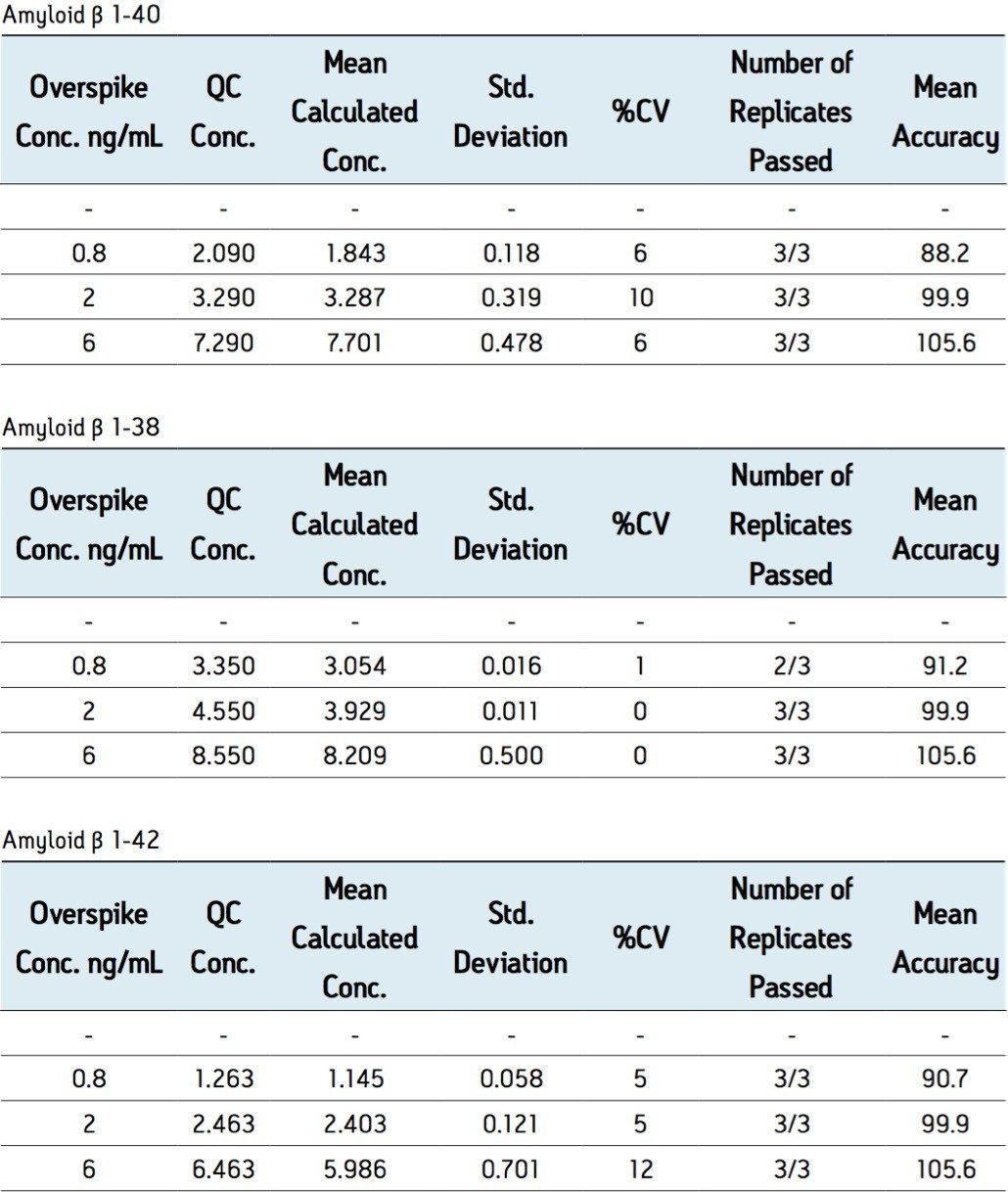
Overspiked QC samples were prepared in 3 sources of pooled human CSF and 1 source of pooled monkey CSF at 0.2, 0.8, 2, and 6 ng/mL. Accuracy and precision values met the regulatory criteria for LC-MS/MS assays. Representative results from QC sample analysis are shown in Table 3.
720003682, October 2010