
This application note describes the analysis of paper and board food packaging extracts. This method enables confident identification of previously unknown compounds with the potential to migrate into foodstuffs.
Recycling paper and board has clear benefits to the environment, relieving pressures on forestry resources and reducing the amount of waste disposal. Currently, there is limited control over the types of paper and board entering the recycling stream. End use of the recycled paper and board ranges from less demanding applications, such as newspapers and magazines, to cardboard boxes and cartons, and more demanding applications, such as food packaging.
In recent years, there have been issues reported in scientific literature and in the media relating to the use of recycled paper and board in food packaging. Contaminants associated with recycled paper and board have been detected in food. Mineral hydrocarbons have been found from inks used to print newspapers and magazines,1-2 as well as phthalates, such as diisobutyl phthalate from adhesives in catalogues and brochures,3 and photoinitiators and other components from printing on the external surface of the paper and board.4 All of these chemical types have been shown to persist after passing through the recycling process.
This study is part of a larger project investigating suitable sources of paper and board for use in recycled food packaging.5 Four different paper sources (plain white printer paper, newspapers and magazines, corrugated cardboard, and food packaging) have been examined and potential contaminants identified. UltraPerformance LC with high resolution mass spectrometric detection (UPLC/HR-MS) has been shown to be a useful tool to aid with identification of unknown compounds in the area of food contact materials and beyond.6 The accurate mass, isotope patterns, and fragmentation information (if present) allow predictions of elemental composition which can be compared to a database of potential structures, if one is available, adding confidence to the identification process. The instrument used must be sufficiently sensitive and accurate to ensure that compounds are confidently identified.
The use of the ACQUITY UPLC System combined with the SYNAPT G2 HDMS and associated software to detect chromatographic peaks, determine accurate mass, and produce elemental composition is described here. A comparison with a user-prepared database containing over 6000 food contact material ingredients and contaminants is described, and, as an example, one of the proposed chemical structures is confirmed using fragmentation information acquired by MSE without the need for authentic standards.
A selection of foodstuffs in paper and board packaging was purchased from a local supermarket. The food was removed from the packaging, cut into small pieces, and mixed well. The samples included breakfast cereals, pasta, frozen fish, cakes, and other baked products.
A portion of the mixed sample (5 g), d10-benzophenone (100 μL of 1 mg/mL) to act as internal standard and ethanol (20 mL) was added to a vial, capped, and shaken overnight. A portion of the ethanol was removed and directly analyzed.
|
System: |
ACQUITY UPLC |
|
Column: |
ACQUITY UPLC HSS T3 (Part No. 176001133) 150 x 2.1 mm, 1.8 μm |
|
Column temp.: |
45 °C |
|
Flow rate: |
0.45 mL/min |
|
Injection volume: |
1 μL |
|
Mobile phase A: |
Water + 0.1% formic acid |
|
Mobile phase B: |
Acetonitrile + 0.1% formic acid |
|
Time(min) |
%A |
%B |
|---|---|---|
|
0.0 |
90 |
10 |
|
15.0 |
0 |
100 |
|
18.0 |
0 |
100 |
|
18.1 |
90 |
10 |
|
20.0 |
90 |
10 |
|
MS System: |
SYNAPT G2 HDMS |
|
Acquisition mode: |
Resolution mode, MSE |
|
Ionization mode: |
Electrospray positive |
|
Mass range measured: |
50 to 1200 Da |
|
Cone voltage: |
25 V |
|
Capillary voltage: |
1.0 kV |
|
Desolvation temp.: |
500 °C |
|
Source temp.: |
120 °C |
|
Collision energy: |
Function 1 CE = 6 eV, Function 2 CE = 15 - 35 eV |
|
Collision gas: |
Argon |
|
Lock mass: |
Leucine enkephalin, m/z 566.2771 |
|
Data management: |
ChromaLynx XS and MassFragment software |
The base peak ion chromatogram (BPI) for the ethanol extract of the pooled food packaging sample is shown in Figure 1.
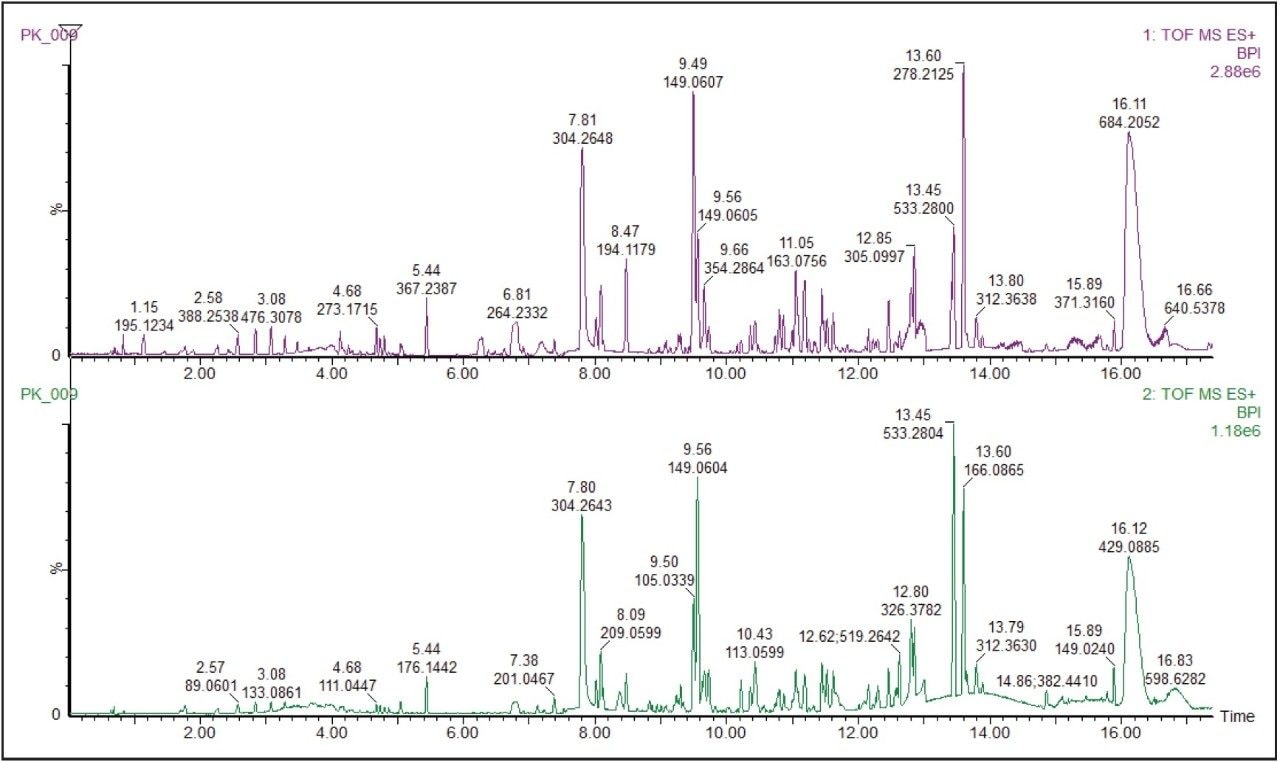
ChromaLynx XS Software deconvolutes chromatograms, detects all chromatographic components present, and produces refined spectra for each identified component. These were processed in the ‘targeted mode’ producing a list of individual peaks that were then compared to a database containing potential structures. The software extracted 1380 individual components, many more than were evident from the TIC, highlighting the power of the software to detect components present at very low concentrations. ChromaLynx XS extracts the exact mass chromatograms of the targeted compounds and confirms their presence or absence.
The user-prepared database contains over 6000 known ingredients, potential contaminants, and reaction and breakdown products in food contact materials. The list contains the compound name and chemical formula that the software will search and report positive hits. Retention time and fragment ion information can also be included in the database, if authentic standards are available and have been analyzed. Figure 2 shows an example of the ChromaLynx XS output including: (A) the TIC, (B) the target list, (C) in particular the extracted ion chromatogram, and (D) associated mass spectrum for the peak at 13.6 minutes, as an example of the completed identification process. Of the 6000 compounds screened in this sample, a total of 45 were identified based on accurate mass. In the absence of analytical standards, these identifications can be supported using the simultaneously acquired fragment information.
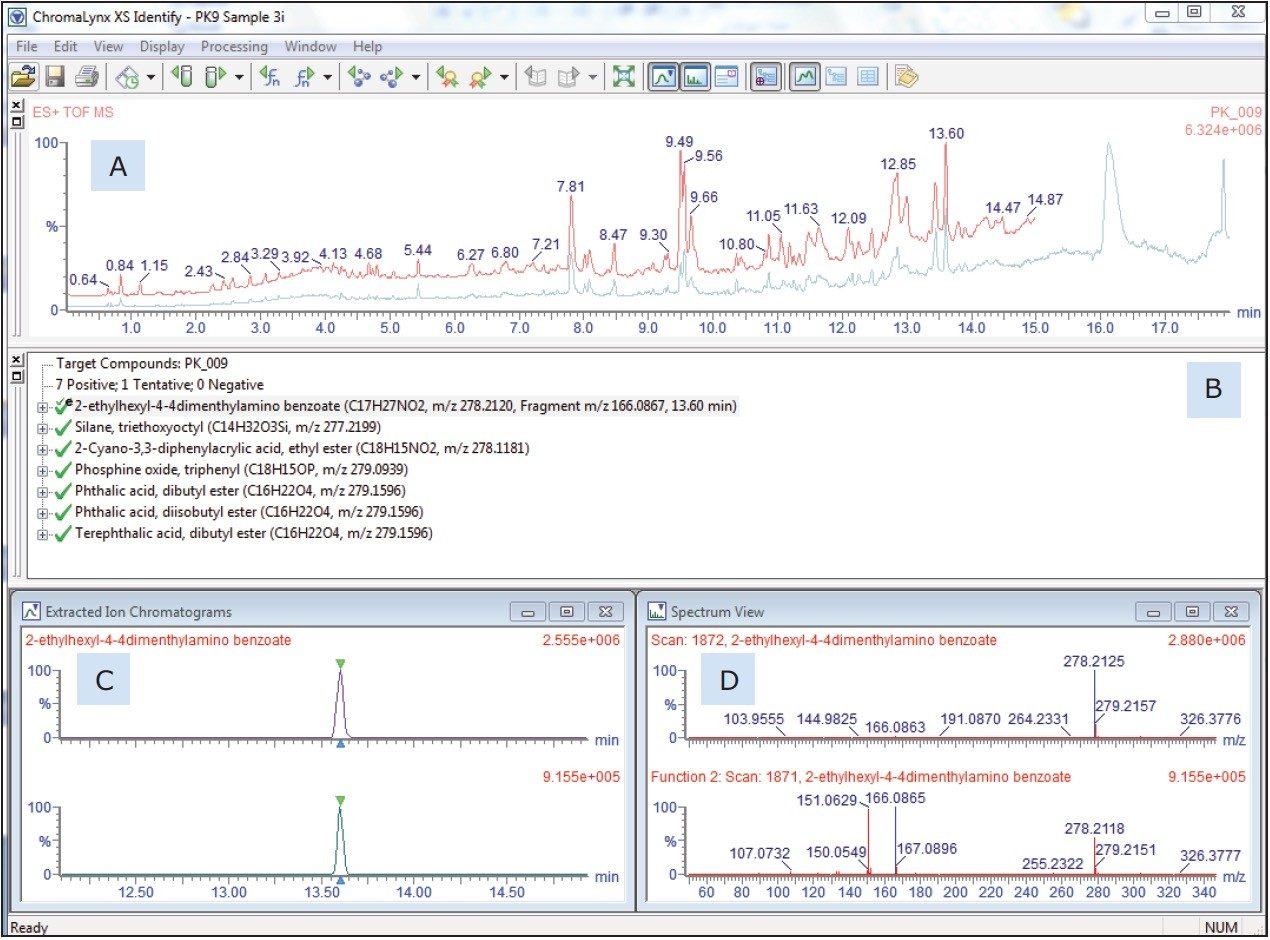
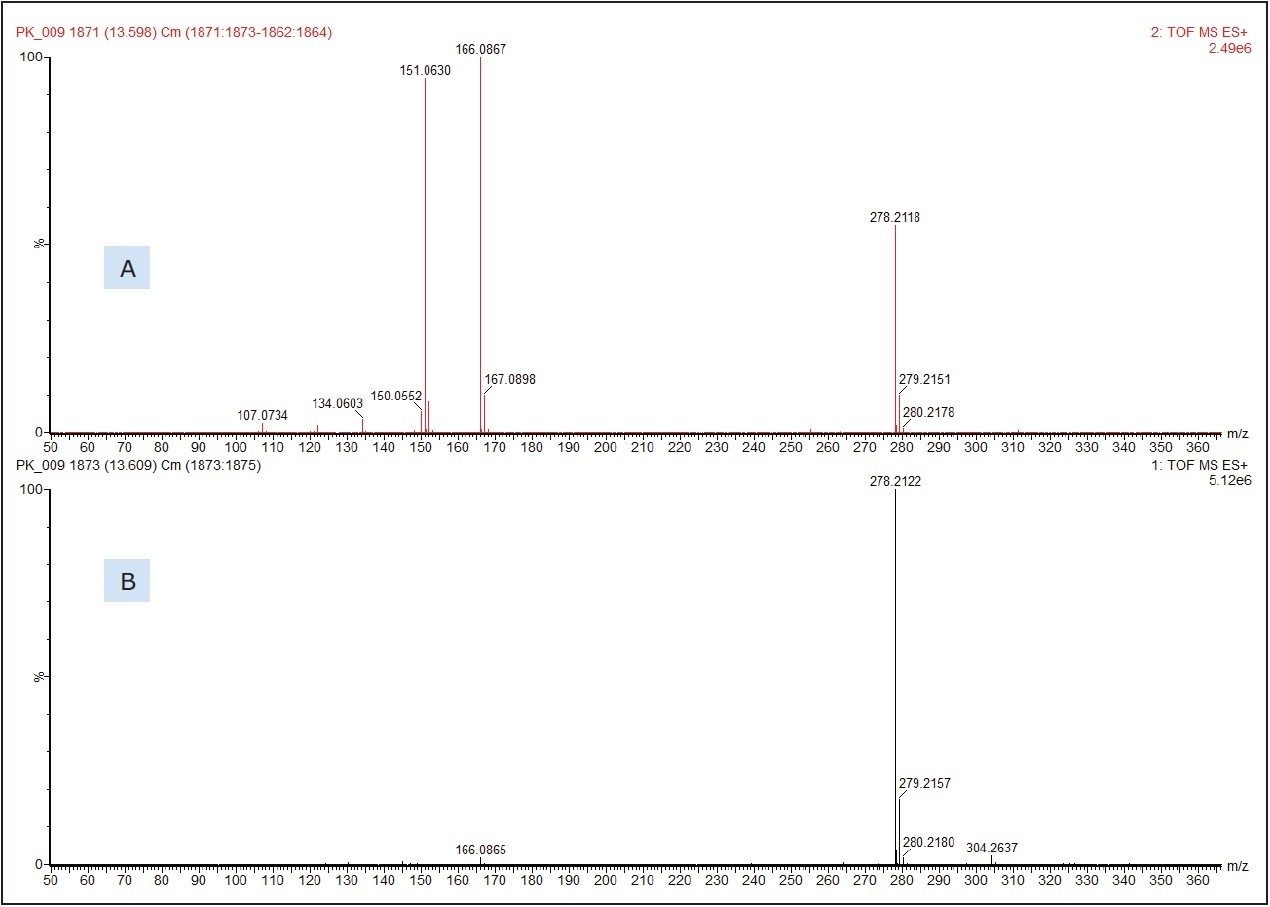
Figure 3 shows the molecular species, [M+H]+ at m/z 278.2122 which produced a chemical formula of C17H27NO2. This gave a database hit of 2-ethylhexyl-4-(dimethylamino)benzoate, which is used as an amine co-initiator in UV-cured inks applied to paper and board substrates. The formula for the [M+H]+ has a theoretical accurate mass of m/z 278.2120, differing by only 0.7 ppm from that measured. No authentic standard of 2-ethylhexyl-4-(dimethylamino)benzoate was analyzed at the same time as the food packaging sample to confirm identification. The SYNAPT G2 HDMS, however, was run in MSE acquisition mode. This allows for the simultaneous collection of both low energy precursor ion ([M+H]+ in this example) and higher energy fragment ion data from a single injection for greater confidence in compound identification.
Figure 3 shows the low and high energy mass spectra with the molecular adduct reducing in intensity at the higher energy, and fragment ions being formed.
Like the molecular species, the accurate mass of the fragment ions can be used to determine potential elemental compositions. These were used in the MassFragment Software to determine likely structures based on the chemical structure of the proposed compound, for example 2-ethylhexyl-4-(dimethylamino)benzoate. The software utilizes systematic bond disconnections and a scoring system dependent on the types of bonds disconnected and the likelihood that this would happen. Inputting information into the program is simple. A .mol file can either be downloaded from ChemSpider online database or be prepared from most common chemical drawing packages, then imported along with the MSE mass spectrum which provides the fragment ions.
The parameters can be adapted depending on the specific needs of the user. The mass window allowance is particularly important with the smaller the range used, the more confidence given to the structural assignment. In this example, a value of +/- 1 mDa was applied. Figure 4 gives the results generated by the software for the peak at 13.6 minutes, proposed to be 2-ethylhexyl-4-(dimethylamino)benzoate.
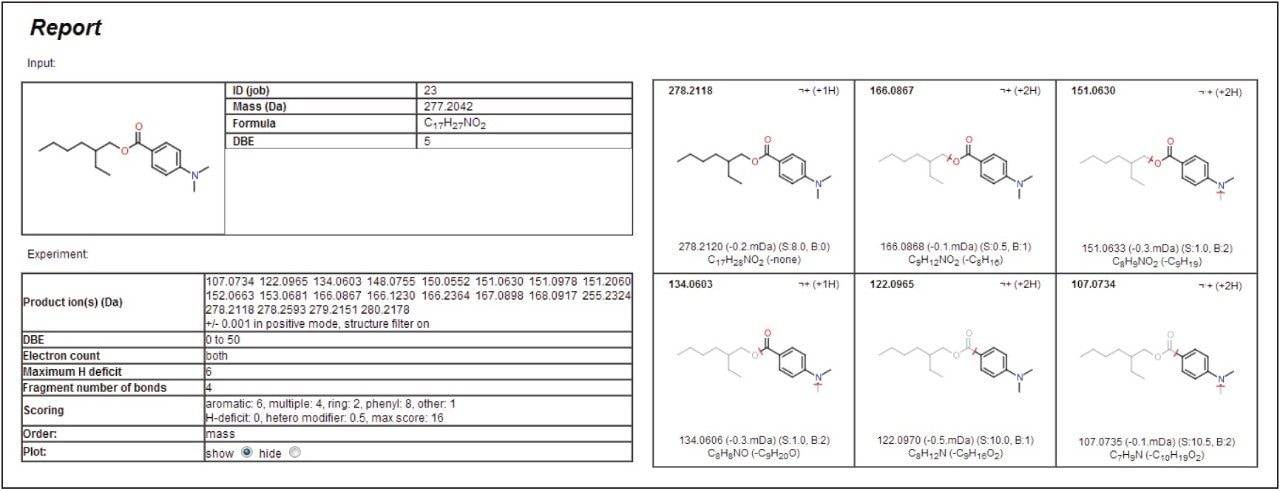
Each of the five fragment ions measured demonstrates that plausible structures have been suggested based on breaking various bonds in the proposed precursor 2-ethylhexyl-4-(dimethylamino)benzoate, increasing confidence to the assignment of this identity to the peak at 13.6 minutes. Figure 5 shows the MSE spectra with annotated MassFragment structures. This compound is most likely derived from the ink applied to the paper and board,7 but compounds of a similar chemical type have been shown to be persistent after the recycling process. Now that the fragments and retention time have been assigned to this compound, they can be fed back into the database for greater confidence in future identifications.

The chromatographic separation, high resolution, and accurate mass capabilities of the ACQUITY UPLC-SYNAPT G2 HDMS System have been used to perform analysis of paper and board food packaging extracts. This enabled confident identification of previously unknown compounds with the potential to migrate into foodstuffs. Both molecular species and fragment ion information collected using MSE acquisition were processed with ChromaLynx XS and MassFragment software, resulting in high levels of confidence in the resulting identifications.
720004591, February 2013