For research use only. Not for use in diagnostic procedures.
This application note demonstrates that the ACQUITY UPLC/Xevo TQ MS System offers lipidomics researchers the opportunity to achieve high performance reverse phase separation of complex lipid mixtures with highly sensitive and accurate detection.
The field of lipidomics is gaining momentum at an incredible pace. The biological knowledge and potential applications are becoming very attractive to those seeking to better understand disease, and for those looking to develop diagnostics. From an analytical science perspective, there is not yet a dominant technique or technology platform, and many researchers have adapted workflows from other life science applications to solve their problems.
In many laboratories, samples are analyzed directly after sample preparation by ESI-MS without separation, which does provide short acquisition times but it can also create problems.
The dynamic range and sensitivity of the experiment is undoubtedly affected by the complexity of the biological sample. Ion suppression from the most abundant analytes and contaminants must be considered and this is compounded when salt and adduct formation are taken into account. These effects combined limit the detection and quantification of isobaric and low abundance lipid species.
Specificity and quantitation accuracy may also be affected when dealing with lipids in a non-separated mixture. Lipids are a diverse class of related species that differ through minor variations in structure. Lipids within the same class are typically distinct from each other by 2 Da, and as such are easily separated; however, inter-class interference is possible.
Monkey plasma was extracted with cold methanol, and the soluble supernatant was dried and re-dissolved in chloroform/methanol (1:1). After adding non-biological lipids for sample normalization (the same lipids used in tuning), the sample was diluted 10x with acetonitrile. Five μL of injected sample was equivalent to 0.14 μL of raw serum and the final solvent composition was injected in acetonitrile/chloroform/methanol (90:5:5). Triplicate injections were analyzed.
|
LC conditions were previously published1,2 |
|
|
LC system: |
Waters ACQUITY UPLC |
|
Column: |
ACQUITY HSS T3, 2.1 x 100 mm, 1.8 mm |
|
Column temp.: |
65 °C |
|
Flow rate: |
500 μL/min |
|
Mobile phase A: |
Acetonitrile/water (40:60) with 10 mM ammonium acetate pH 5.0 |
|
Mobile phase B: |
Acetonitrile/isopropanol (10:90) with 10mM ammonium acetate pH 5.0 |
|
Gradient: |
40% to 100% B/10 min |
|
MS system: |
Xevo TQ MS |
|
Ionization mode: |
Positive/negative switching |
|
Capillary voltage: |
3.2 kV |
|
Desolvation temp.: |
500 °C |
|
Source temp.: |
150 °C |
|
Acquisition method: |
Multiple reaction monitoring (MRM) |
|
Collision cell pressure: |
3.5 x 10-3 mbar |
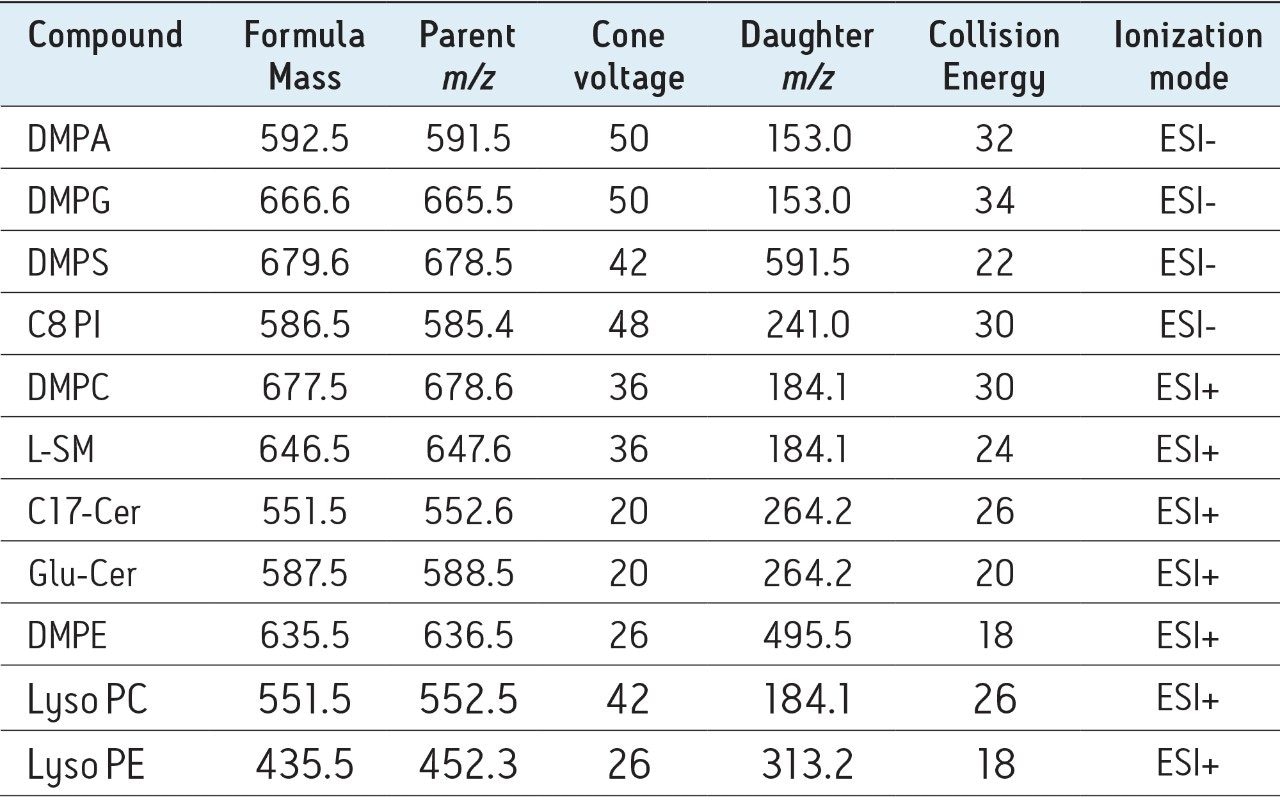
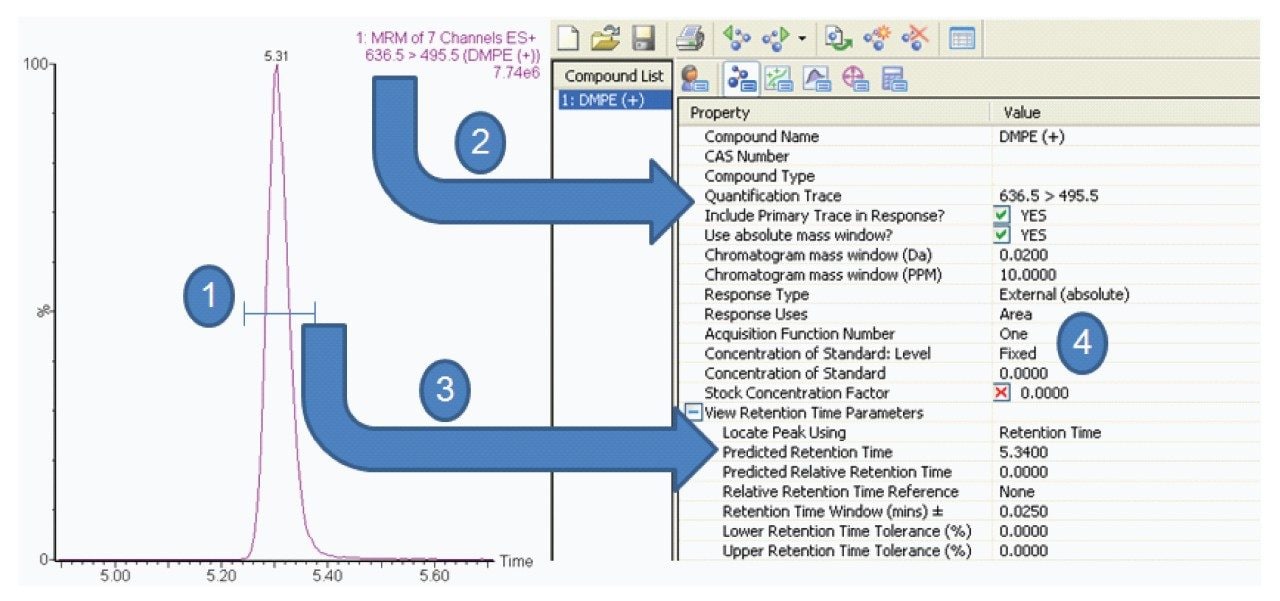
IntelliStart is used to assist the user in the generation of a new analysis, as shown in Figure 1. Assisted MRM tuning for phospholipids was performed for each class with IntelliStart using mixed standards, as shown below in Table 1. For established methods, all of the required experimental parameters including the data analysis template were automatically loaded from Quanpedia.
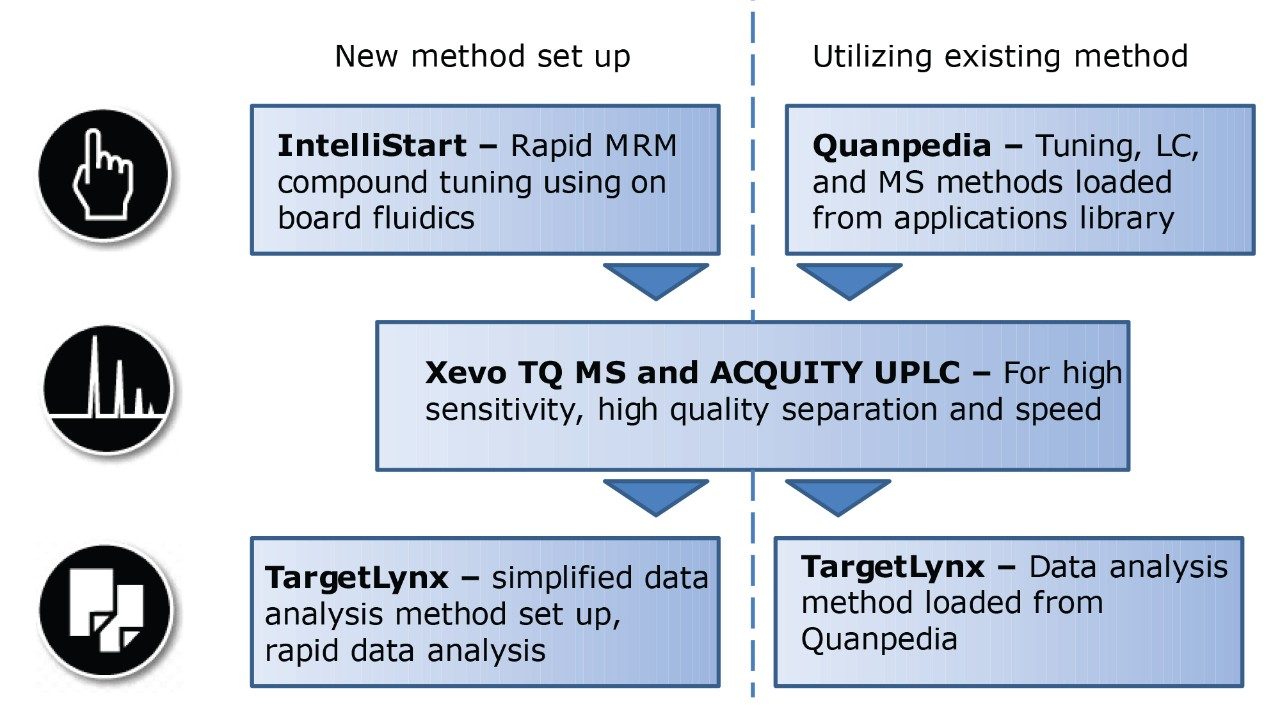
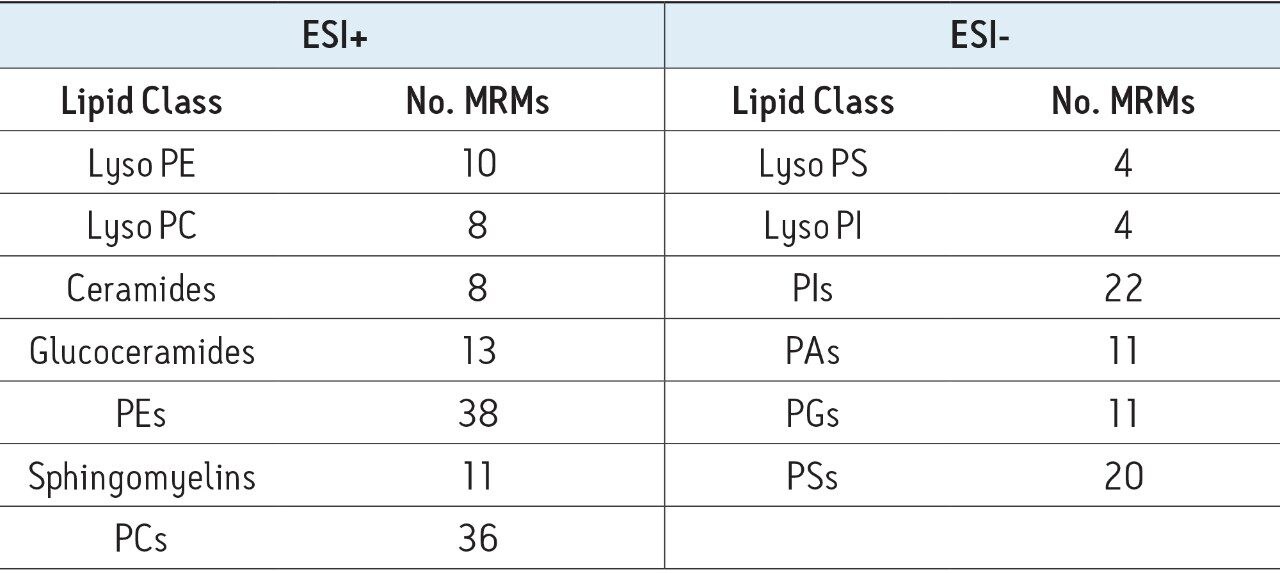
Extracted ion chromatograms for target lipids were easily generated using TargetLynx. Mass chromatograms visualized in the MassLynx chromatogram window can be used to generate quantification methods in TargetLynx, with a single mouse click to transfer retention time, channel, and MRM information. Additional information may be entered to normalize or quantify from an internal or external standard. Once generated, the TargetLynx method can be reused for later experiments.
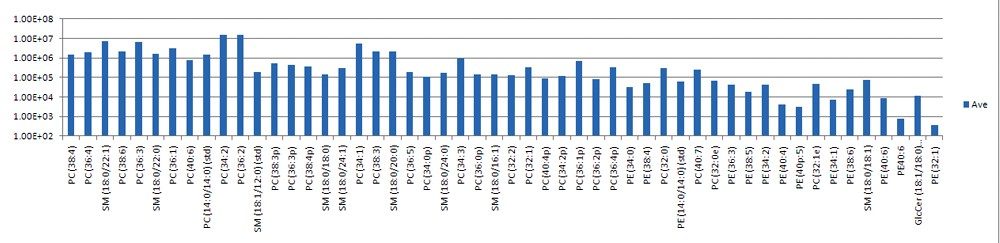
Targeted phospholipids were measured over approximately five orders of dynamic range. Areas under the extracted mass chromatograms were exported from TargetLynx, and selected lipid intensities are shown in Figure 3. The average %RSD between the three replicates for these intensity measurements was under 10%.
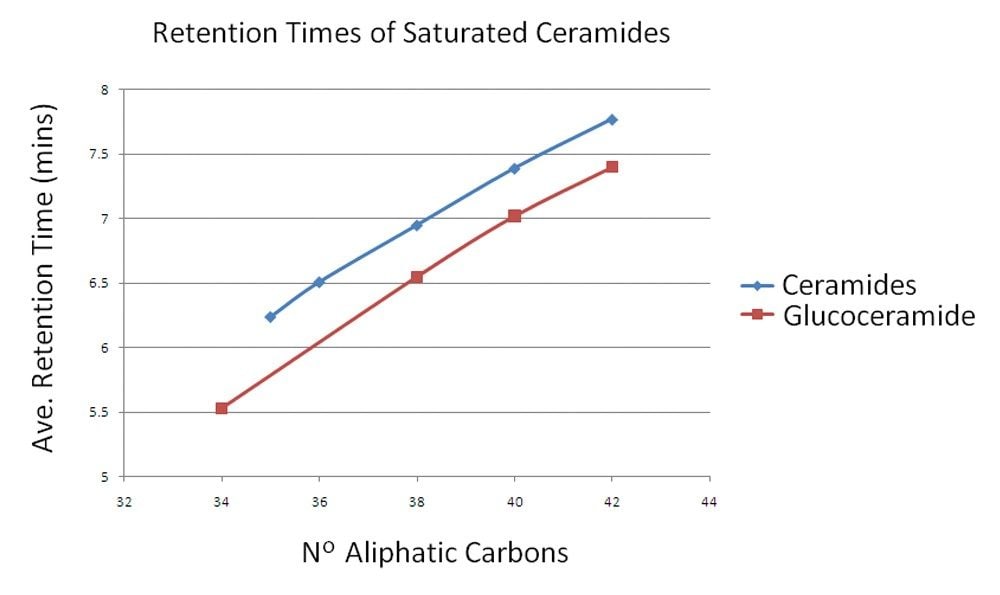
As previously noted1,2 this chromatographic method provides a reliable and reproducible separation of lipids. From this data it is also apparent that lipids in the same class can be separated cleanly and predictably. Figure 4 shows the separation of unsaturated ceramides and glucoceramides as an example. As the aliphatic chain is extended by two carbons, approximately 30 seconds is added to the retention time. Errors in quantification of lipids are often seen in infusion experiments due to overlapping isotopic patterns of lipids that differ by only a single saturated bond. In these cases, the mono-isotopic ion of the higher mass lipid species overlaps with the M+2 isotope of the lower and therefore shows increased abundance for that lipid. In this UPLC method, however, this type of variation in lipids results in significant and predictable chromatographic separation, as shown in Figure 5. In the example shown here, PC 36:1, PC 36:2, PC 36:3, PC 36:4, and PC 36:5 were each separated by approximately 20 s from their closest relative, with the most saturated being the earliest eluting species. In this way, the problem of overlapping isotopic patterns can be overcome.

The ACQUITY UPLC/Xevo TQ MS System offers lipidomics researchers the opportunity to achieve high performance reverse phase separation of complex lipid mixtures with highly sensitive and accurate detection. The reverse phase method uses more stable and less toxic solvents than the usual normal phase separation methods applied to lipidomics, and it is able to differentiate between similar and isotopically interfering species.
In addition to the benefits of separation of similar species, UPLC’s concentration effect and reduction in ion suppression improve the performance dramatically compared to infusion or HPLC experiments. The sensitivity of the technique is apparent as the sample load required by this approach was 1/10th of the sample load that would typically be used in an infusion experiment.
The use of IntelliStart for rapid automated tuning, Quanpedia to generate methods, and TargetLynx for data review provide a simple workflow for efficiently extracting qualitative and quantitative information from large data sets.
720004035, July 2011