
The application of a small negative ion natural products library that comprises 299 analytes is presented and is used to illustrate multi-factor authentication specificity of CCS libraries and their further extension to be used to screen for natural products. The use of LC-MS based methods to profile medicinal plants/herbal remedies and complex samples continues to increase. This approach has become more pivotal in phytochemical makeup profiling but remains a challenge due to sample complexity. Green tea extract has been screened and analyzed. Although the phytochemical makeup of green tea has been extensively profiled by numerous research groups, we report both previously identified and unidentified flavonoid isomers, and provide an illustration of unique TWCCSN2 specific identification profiling.
The application of LC-MS-based methods to profile flavonoid markers in natural products has significantly increased in recent years. The importance of such an analytical approach has become more pivotal, both in species profiling and in consumer product authentication due to the challenges associated with increased sample complexity.1 To date, over 4,000 flavonoids have been identified, possessing diverse pharmacological and biological properties.2 Sample clean-up alone, in the form of solid-phase extraction (SPE), does not alleviate such complexity through extraction selectivity, resulting in sample extracts with non-discriminative elution of structurally related compounds.
Complex sample analysis is a significant challenge, where major/minor component coelution can occur due to insufficient chromatographic peak capacity and/or selectivity. Additionally, subtle changes in chromatographic separation, resulting from sample matrix, column loading, and chromatographic conditions selectivity can result in coelution of isomers and target analytes. Full spectra acquisition qualitative profiling using high resolution ToF-MS (time-of-flight Mass Spectrometry) enables large numbers of compounds to be detected in a single acquisition in conjunction with the ability to perform retrospective data review. Typically, ToF-MS accurate mass specificity is used in combination with time tolerances, isotopic matching, fragment ions/ratios, and response thresholds to screen for target compounds. Despite the selectivity and specificity advantages of high-resolution accurate mass spectrometry including MS/MS, it still can be challenging to determine the identity of analytes. This is especially the case when isomers/structurally related compounds produce the same isomeric product ions, and particularly so with large numbers of co-extracted matrix components.
Waters first developed and commercialized the SYNAPT ion mobility mass spectrometry (IM-MS) platform in 2006, enabling the development and use of mass spectrometry libraries to evolve. Incorporation of IM collision cross section (CCS) values enables identification that is based upon the cumulative metrics of UPLC-IM-MS (ultra-performance liquid chromatography ion mobility mass spectrometry). The IM-based measurement/gas phase separation of compounds is based on size, shape, and charge differentiation prior to the mass analyses.3,4 It provides an added dimension of separation, delivering increased system peak capacity and TWCCSN2 (travelling wave CCS using nitrogen buffer gas) to be used as an additional analyte identification metr
The analytical time scales are UPLC (seconds), ion mobility spectrometry (IMS) (milliseconds (~10 ms)), and time-offlight MS (microseconds). The speed of travelling wave ion mobility separation is compatible with the requirement of high throughput analysis of complex mixtures such as natural products. At acquisition rates of one spectra/second more than 90 analyte data points are readily generated, facilitating flexibility in the complexity of IM experimental design without loss of data integrity. IM small molecule CCS searchable databases have been created across multiple areas of research including natural product isomers (glycosidic catiomers, isomeric disaccharides), natural product screening/metabolism, veterinary drugs, metabolomics, lipids, mycotoxins, steroids, steviol glycosides, and charged isomers.5-20
A strategy to facilitate the efficient and robust generation of mass spectrometry libraries that incorporate precursor/ product ions, adduct ions, retention times, and CCS values has been developed. Briefly, the non-targeted library building strategy utilizes analytical standards that are screened in triplicate, using both positive and negative ion electrospray ionization. Average CCS values (including adducts) and product ions are extracted from UNIFI ion mobility- processed data to produce analyte-specific MS library information.21, 22
The application of a small negative ion natural products library that comprises 299 analytes is presented and is used to illustrate multi-factor authentication specificity of CCS libraries and their further extension to be used to screen for natural products. The use of LC-MS based methods to profile medicinal plants/herbal remedies and complex samples continues to increase. This approach has become more pivotal in phytochemical makeup profiling but remains a challenge due to sample complexity. Green tea extract has been screened and analyzed. Although the phytochemical makeup of green tea has been extensively profiled by numerous research groups, we report both previously identified and unidentified flavonoid isomers, and provide an illustration of unique TWCCSN2 specific identification profiling.23,24
Green tea extract (p/n: 186006962).
33 mg was dissolved in 2 mL of methanol/water 1/3 solution and diluted further, to a working concentration of 8.25 mg/mL for analysis.
|
LC conditions |
|
|
LC system: |
ACQUITY UPLC I-Class PLUS |
|
Detection: |
Ion mobility mass spectrometry |
|
Vials: |
LC-MS Certified Clear Glass 12 × 32 mm Screw Neck Total Recovery Vial, with Cap and Pre-slit PTFE/Silicone Septa, 1 mL volume (p/n: 600000671CV) |
|
Column: |
ACQUITY UPLC BEH C18, 1.7 μm, 100 mm × 2.1 mm, (p/n: 186002346) |
|
Column temp.: |
45 °C |
|
Sample temp.: |
10 °C |
|
Injection volume: |
10 μL |
|
Flow rate: |
0.75 mL/min |
|
Mobile phase A: |
0.1% formic acid in water |
|
Mobile phase B: |
0.1% formic acid in acetonitrile |
|
Gradient: |
0–1 min. isocratic at (98:2.0 (A:B)); 5 min. (95:5); 10 min (80:20); 13 min (70:30); 15 min (20:80); 15.1 min (98:2.0) and 17 min (98:2.0) |
|
MS system: |
SYNAPT G2-Si QTof |
|
Ionization mode: |
ESI |
|
Capillary voltage: |
2.2 kV |
|
Cone voltage: |
30 V |
|
Desolvation temp.: |
600 °C |
|
Source temp.: |
150 °C |
|
Acquisition range: |
50–1200 m/z |
|
Acquisition rate: |
10 spectra s |
|
Lockmass: |
Leucine enkephalin (C28H37N5O7 (m/z 556.2766 +ve/m/z 554.2620 -ve)) |
|
Collision energy: |
HDMSE low collision energy 4 eV and high collision energy ramp (30 to 75 eV) |
|
MS resolution: |
20,000 resolution full width half maximum (FWHM) at 556 m/z |
|
IM resolution: |
≈ 40 Ω/ΔΩ (FWHM) |
|
IMS parameters: |
default IMS screening parameters include T-Wave Velocity Ramp = Start: 1000 m/s End: 300 m/s, T-Wave Pulse Height = 40 V and a gas flow of helium 180 mL and nitrogen 90 mL (buffer gas) for the respective gas cells was used, giving an IM cell pressure of ~3.2 mBar |
|
Calibration: |
IMS/ToF Calibration Kit (p/n: 186008113) |
|
Chromatography software: |
MassLynx v4.1 SCN 916/924 |
|
MS software: |
MassLynx v4.1 SCN 916/924 |
|
Informatics: |
MassLynx data post processed using UNIFI v1.92 |
A negative ion mode mass spectrometry library incorporating accurate mass precursor and product ions retention with times has been developed. CCS values have been generated for precursor ion and adducts. Green tea extract has been screened to assess the robustness of the library produced. The UPLC-MS BPI chromatogram obtained, and the corresponding orthogonal separation presented in Figure 1, have shown where it is evident that chromatographic coeluting analytes are resolved using ion mobility (separation from shape, charge, and mass). In negative ion mode ESI, the data reveal a higher degree of complexity than is observed using positive ion mode. Due to the acidic nature of the flavonoids and glycosides, greater ionisation efficiency is observed (See Waters Corporation application note 720006650EN for more on the positive ion library).
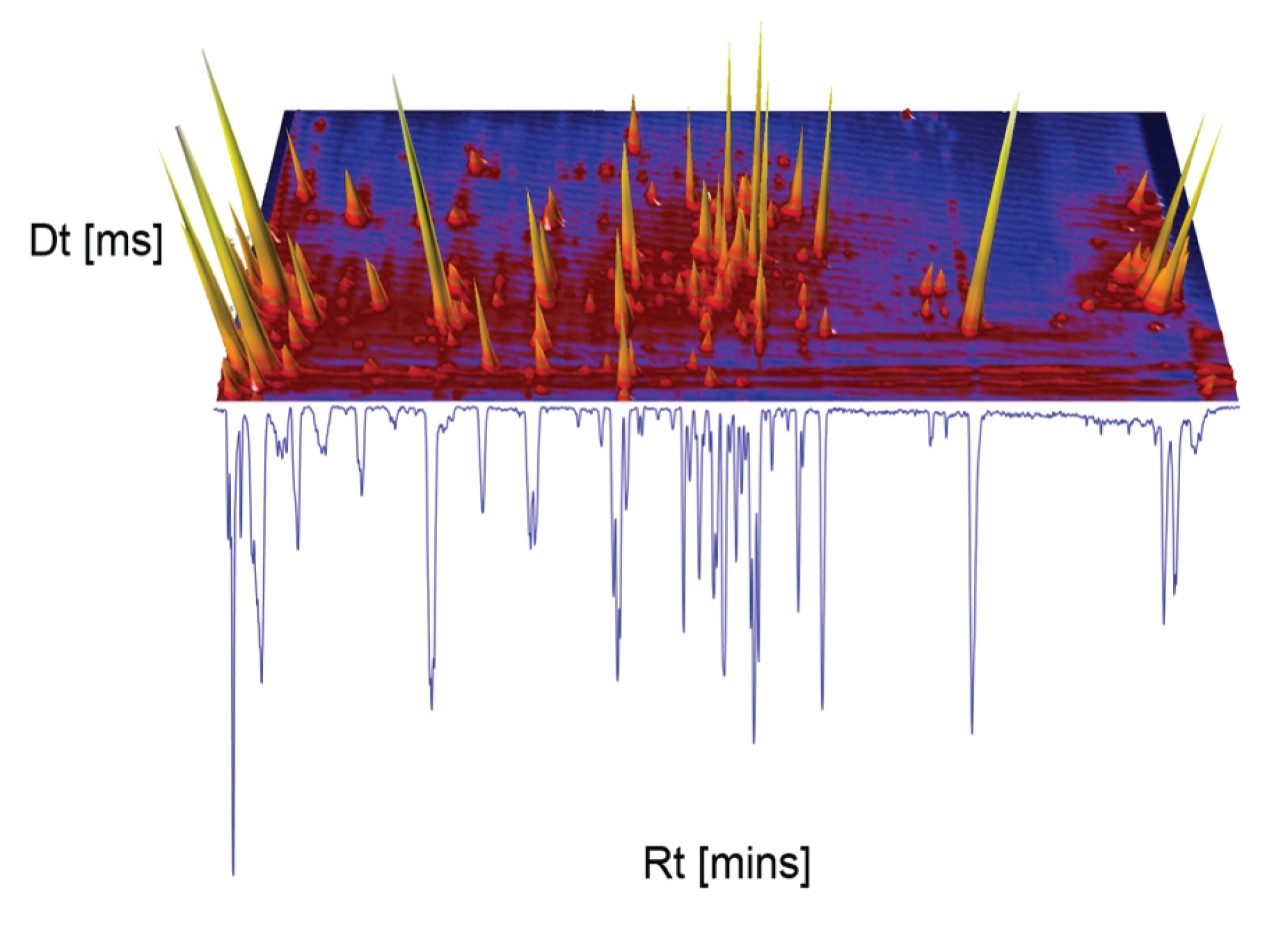
The reversed phase gradient used to generate the retention times of the natural products library constituents was used generate chromatographic separation of the green tea extract constituents. The component summary for the identified analytes using negative electrospray ionisation is shown in Figure 2. Analytes in the following classification groups were identified; amino acid [1], 7-C glycosylated flavones [15], flavanols [12], flavan-3-ols (catechins) [2,4], chlorogenic acids [3], and flavonoids [6-8, 9-17], and flavones [17]. The natural products library generated can be readily extended through further data interrogation and identification of analytes, in natural product extracts. TWCCSN2 specificity enables mass spectrometry libraries to be extended for both knowns and known unknowns.16
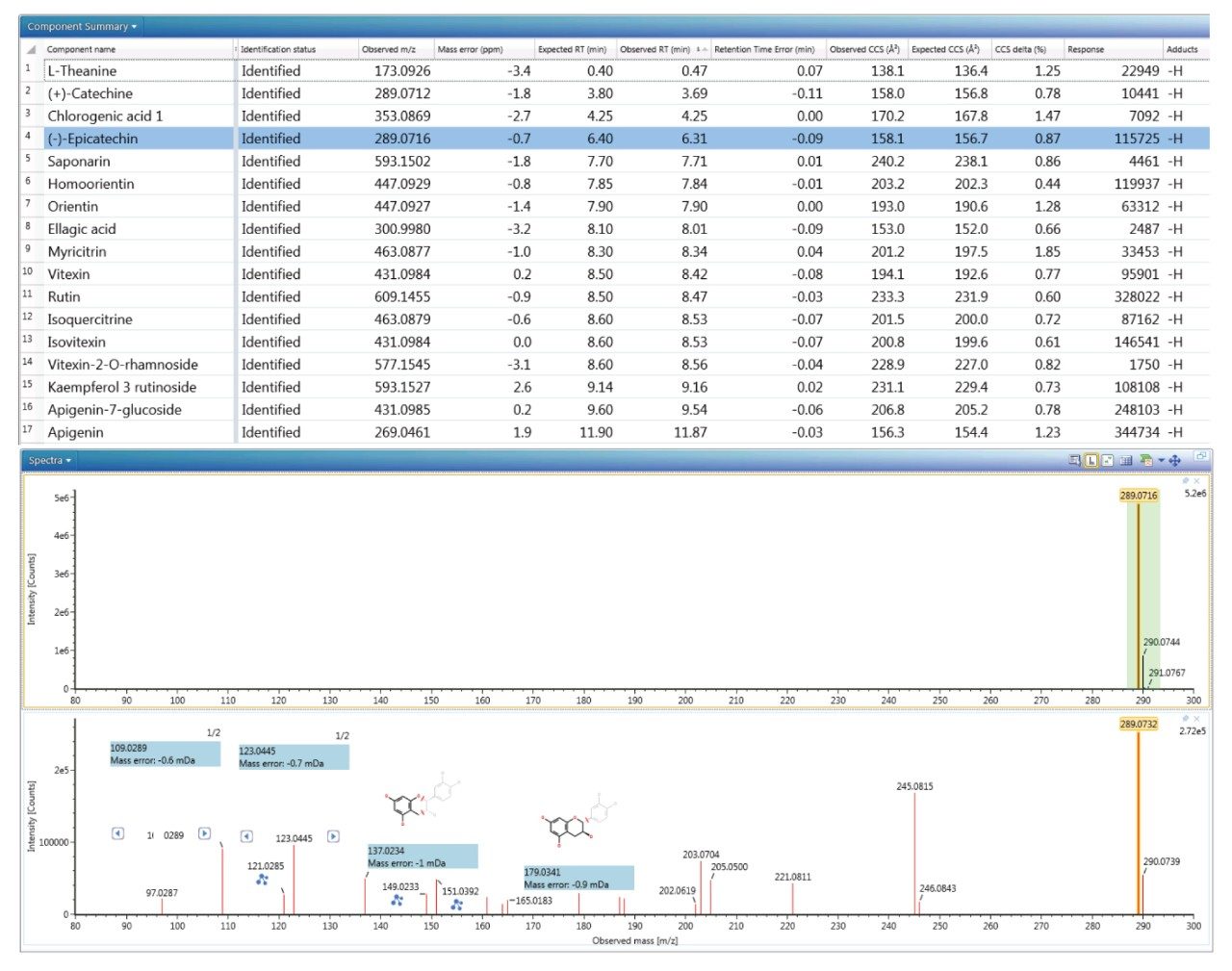
The results shown in Figure 2 illustrate a mass accuracy of the order of 3 ppm and CCS delta of 1%. Moreover, a CCS metric provides an additional identification point for analytes of low abundance where there is insufficient ion intensity for product ion formation. However, the acquisition process is such that precursor and product ions can be generated for all analytes in one single acquisition. More importantly, the product ions generated are highly specific because they are both retention time and drift time aligned, differentiating them from mass spectrometry techniques that produce retention only time-aligned fragment ions. The ion mobility product ion spectrum of epicatechin detected in green tea is presented in the lower pane of Figure 2. Excellent mass accuracy of the product ions was observed (within 1 mDa) when compared to the expected exact mass product ions of epicatechin in the natural products library. Epicatechin was identified by means of seven identification points in total, precursor ion, four product ions, retention time, and CCS (-0.5% delta).
As mentioned previously, to further illustrate the utility, robustness, and confidence that can be gained when using a natural products multi-metric mass spectrometry library, green tea extract was fortified with a series of known analytes (Tashinone IIA, apeginin, apeginin-7-glucoside, orientin, homoorientin, vitexin, isovitexin).22 Although the fortified analytes contained Tashinone A, it was not was not detected as would be expected, since it ionizes in positive ion mode only. All other fortified analytes were detected and identified correctly, further illustrating the robustness of the natural product libraries generated following the developed library building strategy. The 6-C/8-C glycosides (homoorientin/orientin and isovitexin/vitexin) were detected prior to fortification as well, as can be seen in the component summary presented in Figure 2. They have previously been reported to be present in black tea, with just vitexin in green tea. However, using the generated natural products library, both pairs of 6-C/8-C glycosides have been uniquely identified.23,24
When analyzed in negative ion mode, the relative difference characteristic in expected CCS values of the 6-C/8-C glycosides isomer is greater than in positive ion mode (190.6 Å2/202.3 Å2 and 192.6 Å2/199.6 Å2). This occurrence illustrates that, although analytes may ionize in both ionization modes, differing IMS specificity can be observed and determined. Additionally, each of the isomers exhibited characteristic isomeric product ion spectra where distinctive fragment ion ratios further distinguish each respective isomer pair,20 making the identification of these flavonoids in green tea compelling. Reproducibility in the approach discussed herein is shown in Figure 3, where repeat analysis of green tea extract was performed five months after the initial IM screening studies. The long-term robustness of a CCS metric has previously been shown for complex analyses including food commodities (sweeteners and pesticides), animal tissues (veterinary drugs), and natural products.12,16-17
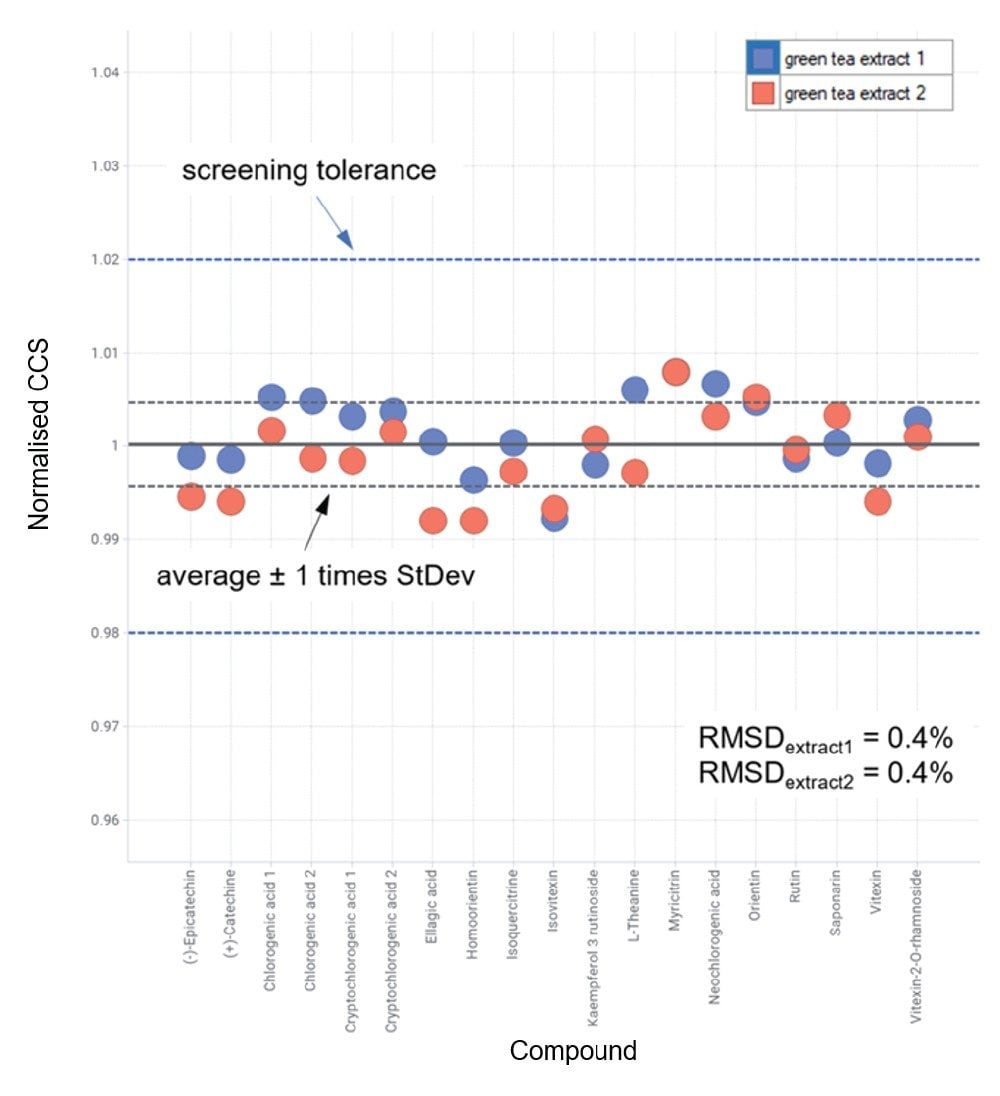
The combined cumulative reproducibility and specificity of a TWCCSN2 metric adds newfound analytical confidence in complex sample analyses to achieve analyte identification of isomers, for example, which as previously stated, can prove to be challenging. To illustrate the latter, in Figure 2, it can be observed that homoorientin is identified at 7.85 minutes and orientin at 7.90 minutes, just 0.05 minutes difference in observed retention time. However, the unique routine incorporation of CCS into the UNIFI mass spectrometry library has enabled the observed differences in CCS in positive and negative ion mode to be utilized. The correct identification assignment has been made routine using unique UNIFI functionality that exploits the highly specific cumulative metrics generated in both ionization modes. IM deconvolution of isomeric product ions produced from isomers eluting in proximity has been performed. Identification of such isomers is highly relevant in many areas of research including medicinal plant profiling where isomeric species can have different phytochemical activity, or in the case of food manufacture, can impact the taste profile of the final product.12
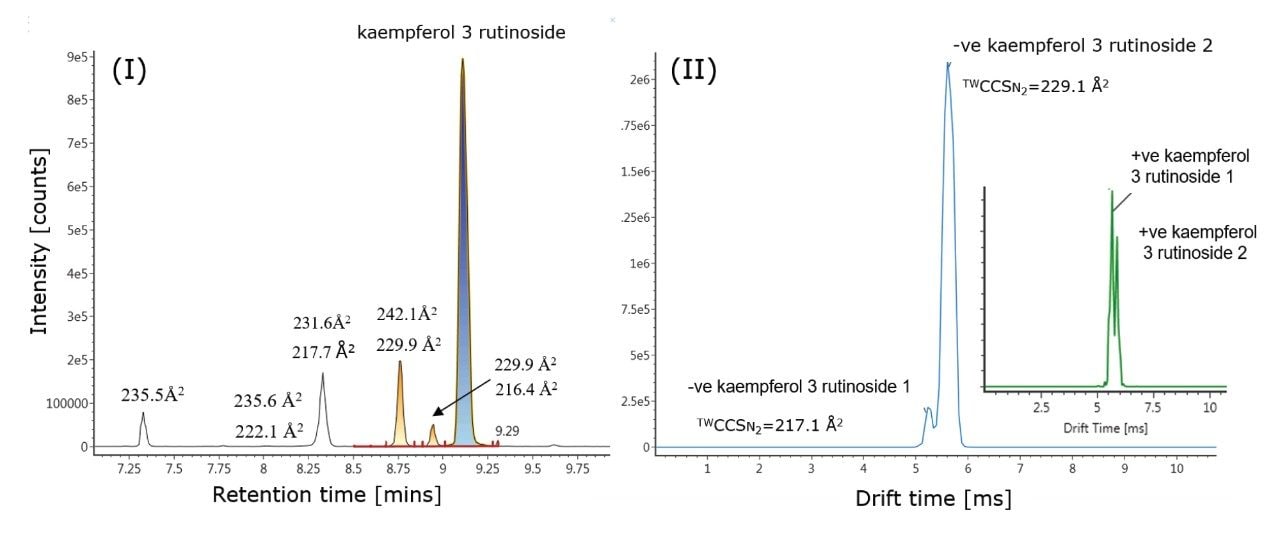
Using analytical standards, it was interesting to observe that for kaempferol 3 rutinoside, two IM- separated species were detected for one chromatographic peak (Rt = 9.12 mins), with determined TWCCSN2 values of 216.1 Å2 and 229.9 Å2. As shown in Figure 4, the two IM-separated species of kaempferol 3 rutinoside were identified in green tea extract with CCS values of 217.1 Å2/229.1 Å2 and respective delta values of 0.5%/-0.4%. In Figure 4(II), it is also shown that in positive ion mode, two IM-separated species were observed for kaempferol 3 rutinoside. In addtion, a potassiated and sodiated adduct were also identified, enabling four positive ion mode TWCCSN2 values to be determined for the same compound in total. Employing an approach to screen using positive and negative ion mode data, a TWCCSN2 fingerprint comprising six CCS values in total was obtained (see Table 1). In addition to retention time, two precursor ion accurate mass measurements and four IM product ion spectra were detected. As seen from the results shown in Figure 5, in negative ion mode, the charged isomer species of kaempferol 3 rutinoside also produces distinctive product ion spectra, which may result from the occurrence of two charge location sites or two conformations.

![IM resolved [M-H]- precursor ion and distinctive product ion spectra](/content/dam/waters/en/app-notes/2020/720006791/720006791en-f5.jpg.82.resize/img.jpg)
Highly specific multi-factor authentication data can be generated using IM screening, in the case of rutin one, IM species is observed in negative ion mode and two in positive ion mode (Waters Corporation Application Note 720006650EN). The advantage of IM screening of complex samples is further illustrated in Figure 4 (I) and Table 2. From the extracted exact mass chromatogram obtained for kaempferol 3 rutinoside, series of additional isomeric species are observed. The corresponding obtained TWCCSN2 values enabled a known unknown TWCCSN2 profile to be generated. Along with kaempferol 3 rutinoside, five of the six isomeric species exhibited two IM charged isomers. Accurate mass measurement alone would not allow such specificity to be discovered and utilized This research further illustrates the unparalleled specificity that can be achieved using UPLC-IM-MS. In addition, IM provides enhanced peak capacity to resolve complex samples, facilitating the acquisition of the non-targeted retention/drift time aligned precursor/product ions of all analytes detected in a single acquisition.
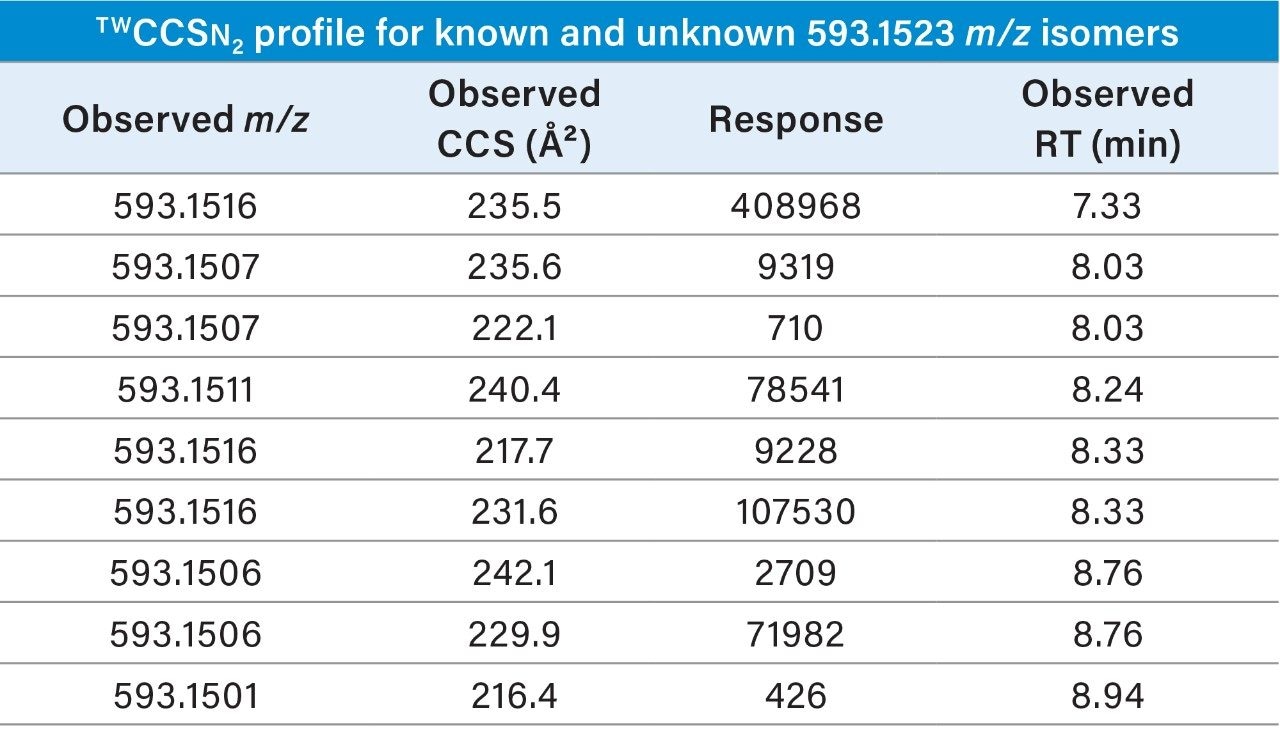
The additional specificity afforded by IM enables flexibility in chromatographic gradients employed and the incorporation into UPLC-IM-MS libraries can potentially reduce the reliance on injecting expensive natural product standards to determine isomer retention times. Moreover, the specificity can aid speed of method development and data interrogation, thereby providing time and cost efficiency. Importantly, the enhanced peak capacity of UPLC-IM provides potential to reveal and gain greater understanding of known phytochemical activity produced by natural product extracts (nutraceuticals and traditional Chinese medicines).
720006791, March 2020