For research use only. Not for use in diagnostic procedures.
This is an Application Brief and does not contain a detailed Experimental section.
This study demonstrates the benefit, reproducibility and applicability of using SONAR on the Waters Xevo G2-XS QTof System for the analysis of biological fluid(s) for a large cohort metabolic profiling study.
Biomedical research and epidemiological studies often consist of large cohorts (>1000 samples) to allow for more statistically relevant findings. The LC-MS system used to perform these analyses needs to deliver a high degree of robustness and reproducibility. The system must demonstrate stability over the entire analysis to ensure potentially subtle biologically relevant features can clearly be detected and identified without batch effects adding to the data complexity.
The statistical analysis tools employed in metabolic profiling, such as principal components analysis, partial least square discriminate analysis, and orthogonal partial least squares discriminant analysis, rely on consistent mass accuracy and reliable concentration assessment for each component in the derived data.
Once features of interest or variance between the subject groups have been detected and proven statistically relevant, a confident identification is required. For this: clear, strong, and accurate fragmentation spectra is critical.
To demonstrate the suitability of the Xevo G2-XS QTof System, operating in SONAR mode, for the analysis of biological fluids for large cohort studies, the system was challenged with the continuous analysis of 900 injections of human urine. A bulk analytical sample was prepared, and non-endogenous compounds were spiked into the sample to act as reference markers.
The analysis consisted of reversed-phase chromatography employing an ACQUITY UPLC HSS T3 Column (1.8 µm, 2.1 x 100 mm). The samples were eluted with an aqueous formic acid – acetonitrile gradient over eight minutes, followed by a high organic wash and suitable re equilibration step at a flow rate of 600 µL/min. The column effluent was analyzed using a Xevo G2-XS QTof Mass Spectrometer operating in SONAR positive ion electrospray mode. The mass spectrometer was operated in sensitivity mode with the synthetic peptide leucine enkephalin employed as the lock mass reference.
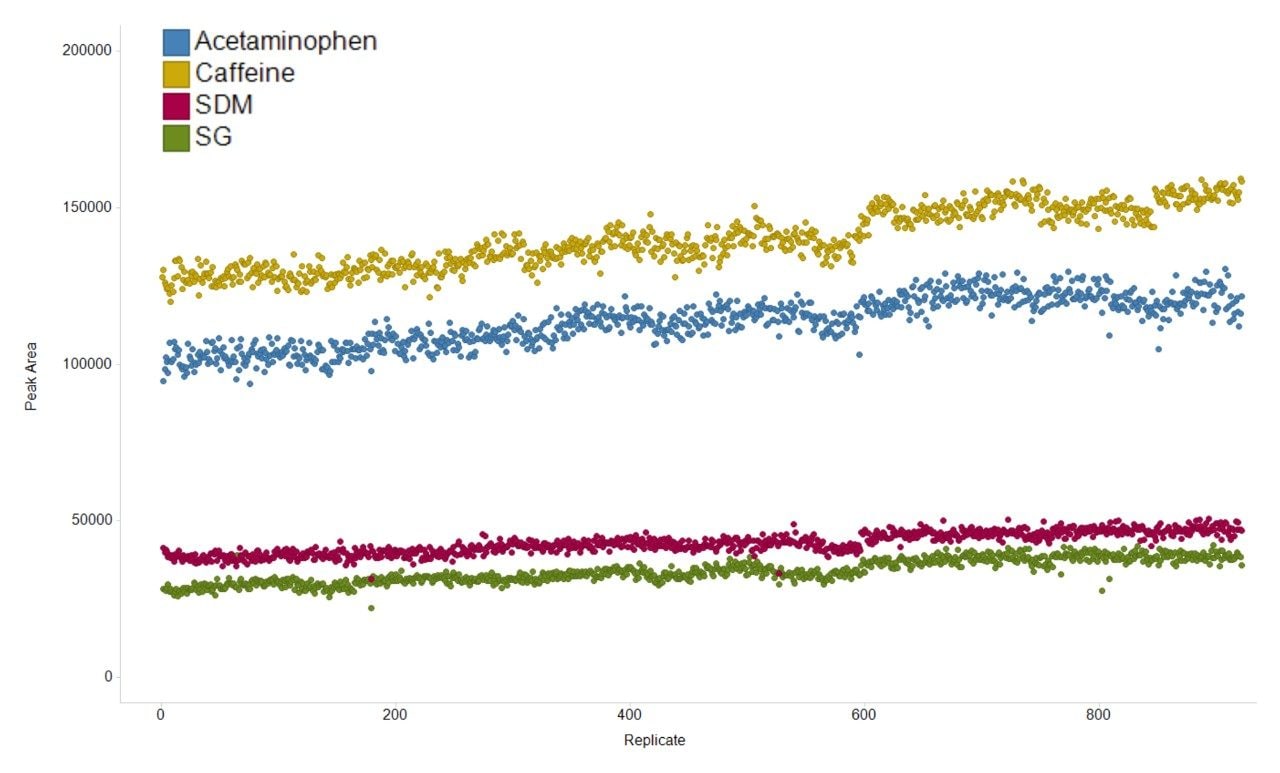
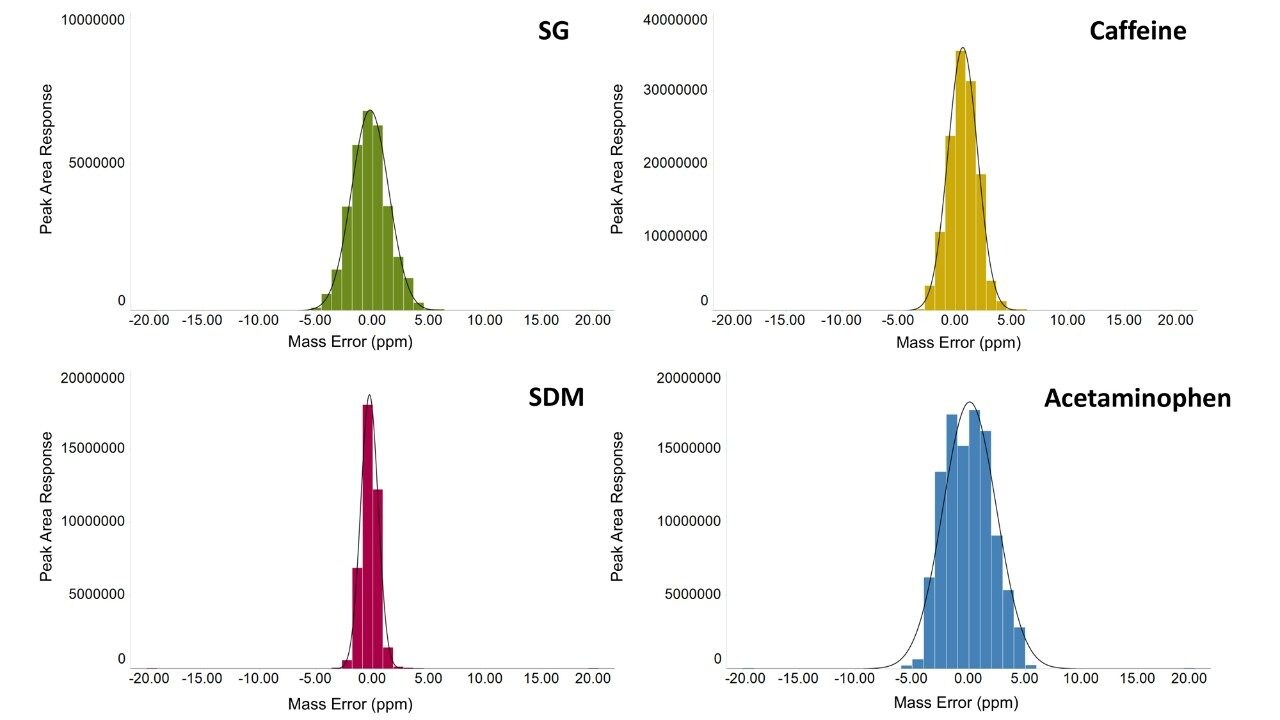
The variation in the mass spectrometry peak area response over the duration of the analytical process for four reference compounds spiked into human urine is shown in Figure 1. The coefficient of variation in response for all 900 injections ranged from 6.7% to 11.1% with a mean variation of 6.4%, with no normalization applied. The mass accuracy, in terms of ppm error, for every injection is shown in Figure 2. The ppm variance of the spiked standards, sample to sample is shown to fall within a 5 ppm (compound dependent variance of +/- 0.0009 to 0.001 Da) tolerance range.
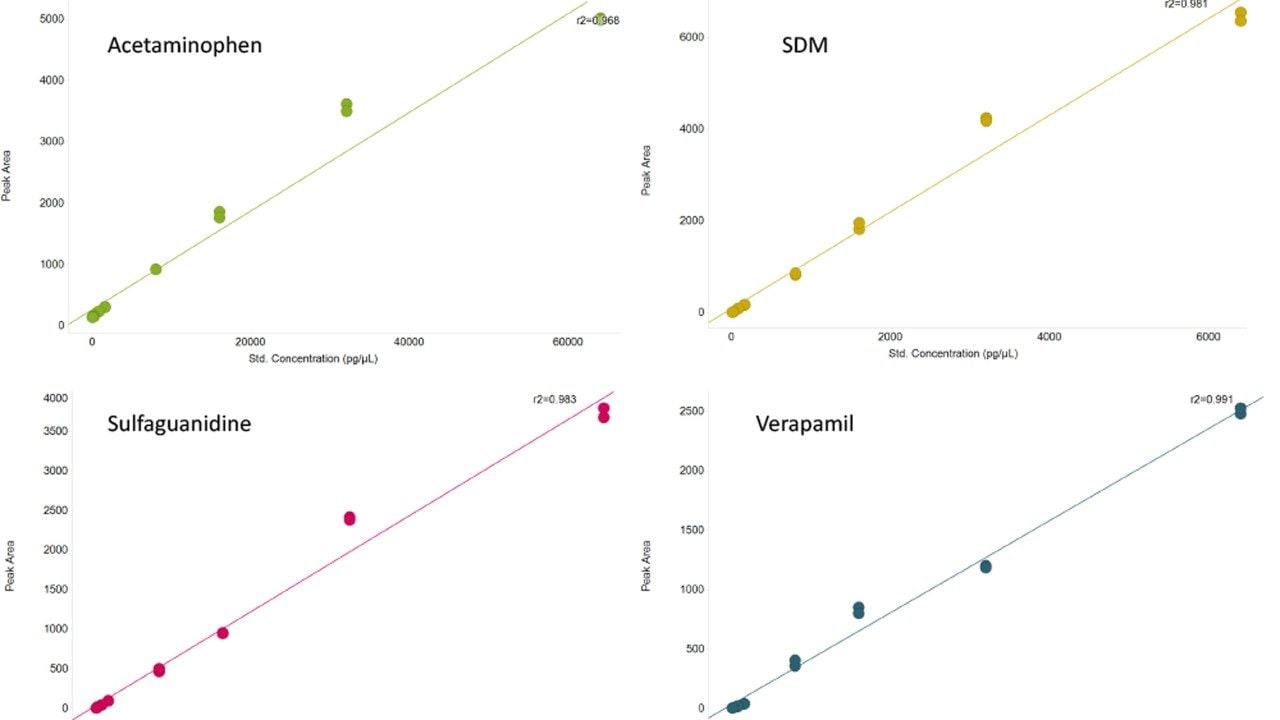
By their nature, biological samples are extremely varied, in particular when individuals may be stressed (e.g., by a disease), leading to metabolites becoming under or over expressed. Measurement of these expression changes requires data to be collected over a suitable dynamic range. To assess this, the linear dynamic range for a number of spiked compounds in matrix were determined (Figure 3). A review of the data demonstrates that the LC-MS system achieves a dynamic range of three orders of magnitude, with lower and upper limits of detection ranging between 64 pg and 32 ng of material injected for acetaminophen or 1.28 pg and 1.28 ng for verapamil, the other spiked compounds are between these concentration ranges.
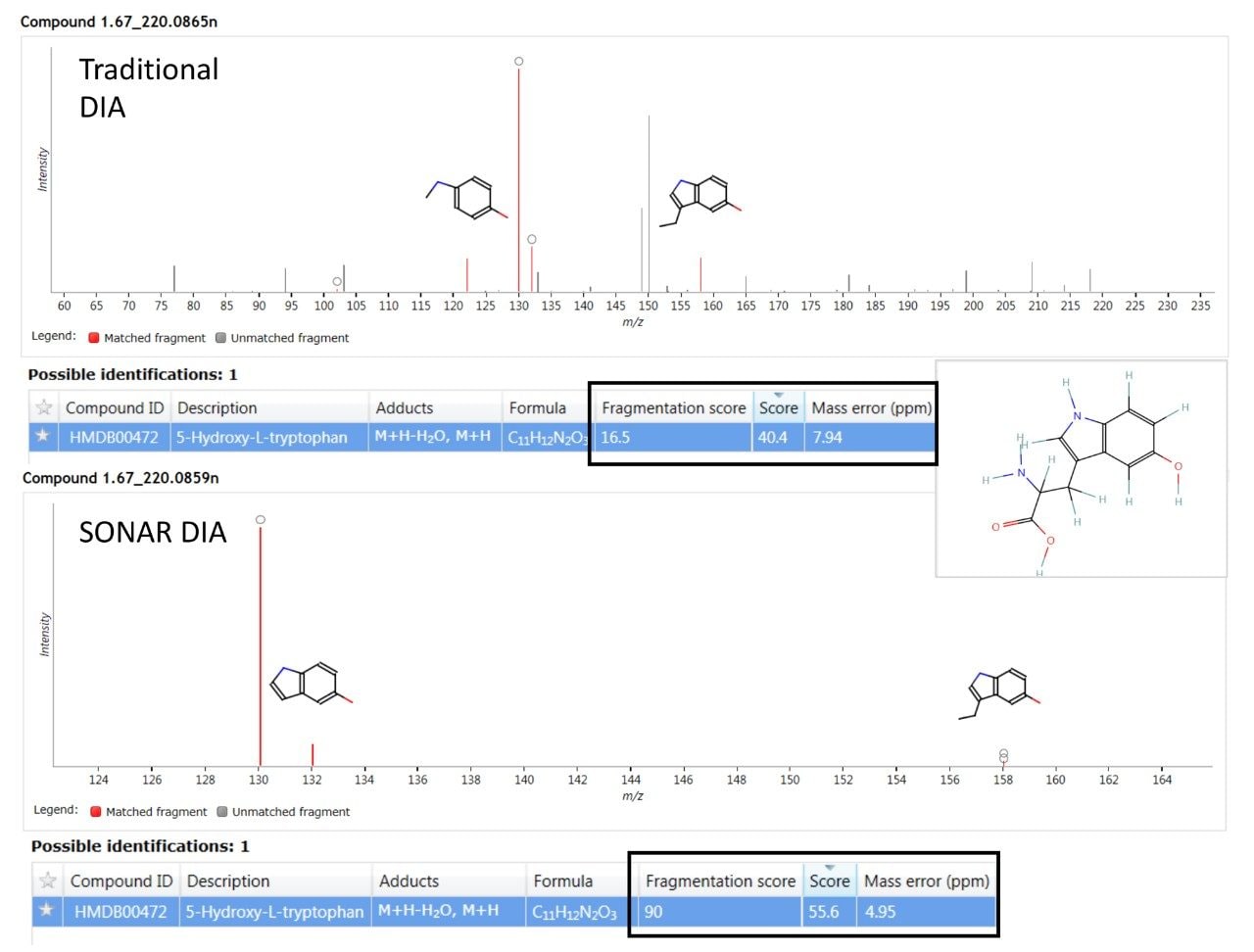
A major benefit of SONAR for the analysis of complex biological matrices is the ability to produce clean, precise fragmentation data for individual precursor ions. The two spectra shown in Figure 4 compares the spectral quality of fragmentation data obtained from a full scan DIA experiment versus the same sample (human urine) acquired using a SONAR enabled DIA analysis. The SONAR spectra show a higher specificity when compared to the conventional DIA experiment and thereby provides increased confidence for compound identification through database searching. Consequently, the results from database searching provided a compound identification for 5-hydroxy-L-tryptophan, with the SONAR spectral clarity resulting in a fragmentation matching score of 90.0 with the conventional DIA method scoring at 16.5. Furthermore, the SONAR acquisition also resulted in an improved database score (55.6) based on the detected nominal mass and that from the database searched against.
This study demonstrated the applicability of SONAR for large scale metabolomic studies. The system showed excellent peak area response reproducibility with a mean variation of 6.4% over 900 consecutive injections, amounting to seven days of continuous analysis time.
The mass accuracy demonstrated by the system shows a gaussian distribution with a maximum ppm error of +/- 5 ppm.
The linear dynamic range was assessed using the spiked compounds into matrix (urine) and this shows linearity over three orders of magnitude with the amount of each compound analyzed in the pg to ng range.
The fragmentation spectra quality using SONAR compared with full scan DIA is significantly improved, with no fragment ions from co-eluting compounds observed for SONAR acquired data and a much improved fragmentation score generated within Progenesis QI.
720006565, May 2019