This application note evaluates HILIC SPE sample preparation to ensure quantitative recovery of both unlabeled and labeled N-glycans.
More than half of all proteins are estimated to be glycosylated.1,2 This posttranslational modification, involving the attachment of oligosaccharides, plays a very significant role in many biological processes.3 Therapeutic antibodies are a salient example of a set of proteins affected by glycosylation, given that their efficacy and immunogenicity can be considerably attenuated by changes in their glycan profile. Glycan profiles of therapeutic antibodies are often, therefore, a critical quality attribute (CQA) that must be assessed during cell line selection and monitored during development and batch releases.
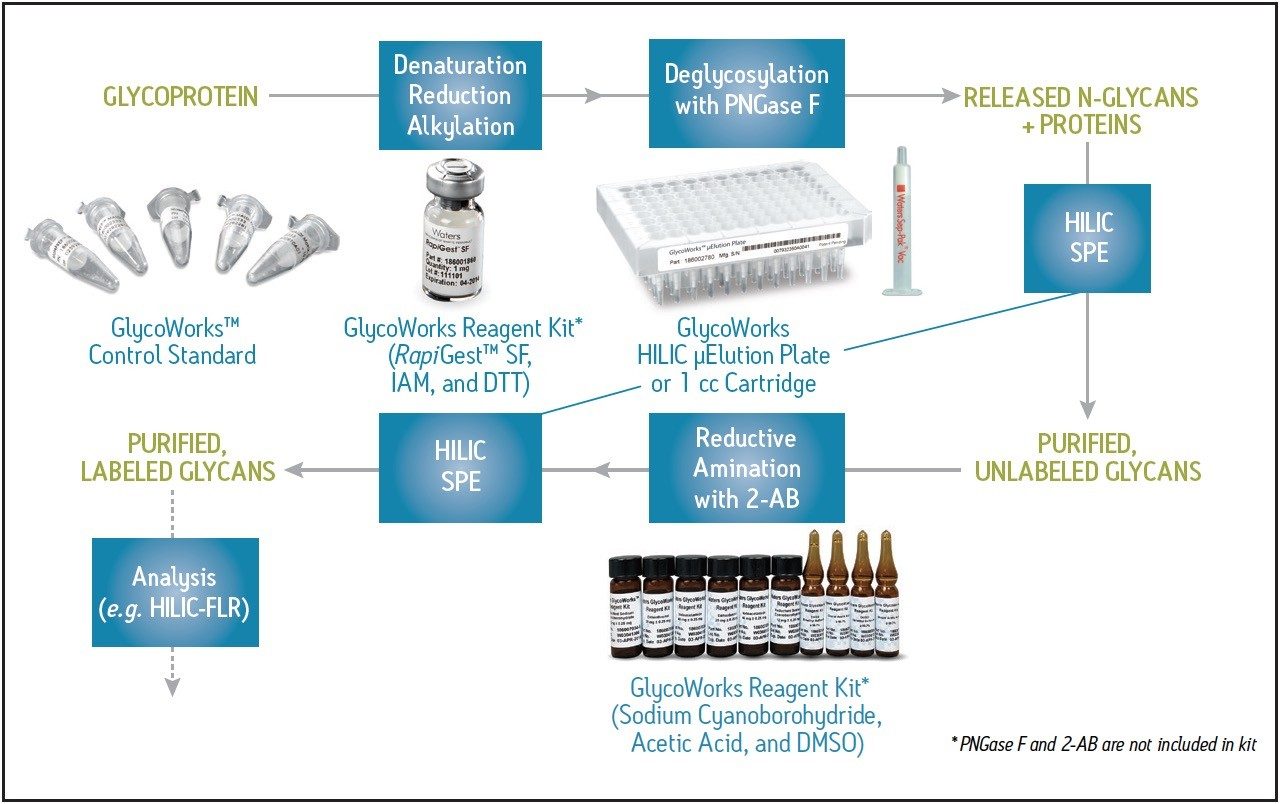
A highly effective analysis platform for evaluating N-glycans from glycoproteins involves the release of glycans by PNGase F, their labeling with fluorescently active 2-aminobenzamide (2-AB), subsequent separation by hydrophilic interaction chromatography (HILIC), and detection by fluorescence (FLR), as shown in Figure 1.3-10
For the labeled oligosaccharide recovery studies, the Glycan Performance Test Standard (p/n 186006349) was mixed with 2-AB labeled trisialylated A3 (ProZyme) in water to make a solution of 3 pmol/μL. Aliquots (10 μL) of this mixture were diluted with 15 μL of acetonitrile (ACN) to make control samples. Aliquots (10 μL) were also dried under vacuum to prepare lyophilization control samples. In addition, 10-μL aliquots were processed by HILIC SPE according to the protocol found in the GlycoWorks High-throughput Sample Preparation Kit Care and Use Manual (p/n 715004079). Various eluents were studied and are noted in Figures 3 through 6. Dried glycans were reconstituted in 10 μL of water and 15 μL of ACN prior to injection.
For the unlabeled oligosaccharide recovery studies, unlabeled Man5 and trisialylated A3, obtained from ProZyme, were reconstituted in water, and mixed to equal molarity (6.7 μM). Aliquots (6 μL) of this mixture were diluted with 6 μL of ACN to make control samples. Aliquots (6 μL) were also dried under vacuum to prepare lyophilization controls. In addition, 6 μL aliquots were processed by HILIC SPE, according to the protocol found in the GlycoWorks High-throughput Sample Preparation Kit Care and Use Manual. Elution was performed with an eluent comprised of 100 mM ammonium acetate (NH4OAc) in 5% ACN (pH 7). Dried glycans were reconstituted in 6 μL of water and 6 μL of ACN prior to injection.
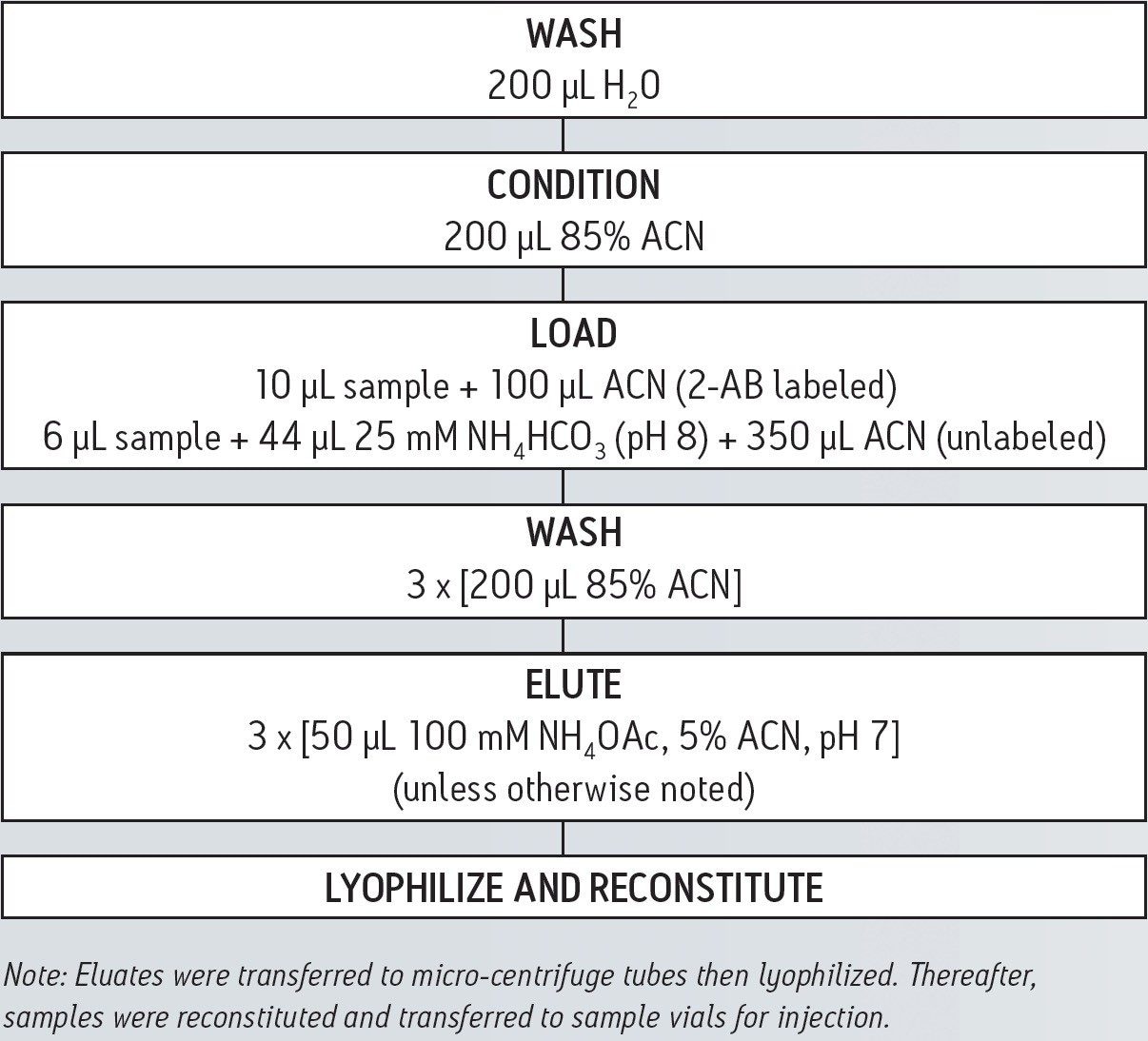
A schematic for the HILIC SPE steps used in this study is shown below:
The procedure for preparing samples for this analysis can be complicated. GlycoWorks products help make the workflow more straightforward by bringing together many of the needed consumables. Moreover, GlycoWorks products provide a solution to the cleanup steps that are needed throughout the process of preparing labeled glycans for analysis. In particular, HILIC solid-phase extraction (SPE)11,12 has been developed to purify released glycans from proteins and buffer/formulation constituents, which can disrupt derivatization. HILIC SPE can also purify labeled glycans after derivatization from excess reagents, which can potentially interfere with downstream chromatography, reduce the lifetime of a column, and thereby impair method robustness.
This application note evaluates HILIC SPE sample preparation to ensure quantitative recovery of both unlabeled and labeled N-glycans. A test mixture, including a complex array of 2-AB labeled human IgG glycans spiked with both high mannose and trisialylated glycans, was used to interrogate and optimize SPE recoveries as well as study the robustness of optimized elution conditions. In addition, an LC-MS assay was employed to demonstrate the quantitative recovery of unlabeled glycans during HILIC SPE with the optimized conditions.
|
LC conditions |
|
|---|---|
|
System: |
ACQUITY UPLC H-Class Bio with a 20-cm Column Heater |
|
Detection: |
Waters ACQUITY UPLC FLR Detector |
|
Excitation: |
330 nm |
|
Emission: |
420 nm |
|
Scan rate: |
10 Hz |
|
Time constant: |
0.2 s |
|
Gain: |
1.00 |
|
Column: |
ACQUITY UPLC GST Amide (BEH Glycan), 1.7 μm, 2.1 x 150 mm (p/n 186004742) |
|
Column temp.: |
60 °C |
|
Sample temp.: |
15 °C |
|
Injection volume: |
2.5 μL (HILIC-FLR), 10 μL (HILIC-MS) |
|
Flow rate: |
0.5 mL/min (0.25 mL/min for the highly aqueous regeneration step in the gradient) |
|
Mobile phase A: |
100 mM Ammonium formate, pH 4.4 |
|
Mobile phase B: |
Acetonitrile (ACN) |
|
Sample collection plate: |
1 mL Round Well Collection Plate (p/n 186002481) |
|
Vials: |
LCGC Certified Clear Glass 12 x 32 mm Screw Neck Qsert Vial (p/n 186001126C) |
|
Time(min) |
%A |
%B |
Flow rate(mL/min) |
|---|---|---|---|
|
0.0 |
22.0 |
78.0 |
0.5 |
|
38.5 |
44.1 |
55.9 |
0.5 |
|
39.5 |
80.0 |
20.0 |
0.3 |
|
44.5 |
80.0 |
20.0 |
0.3 |
|
46.5 |
22.0 |
78.0 |
0.5 |
|
50.0 |
22.0 |
78.0 |
0.5 |
|
Time(min) |
%A |
%B |
Flow rate(mL/min) |
|---|---|---|---|
|
0.00 |
27.9 |
72.1 |
0.5 |
|
19.25 |
50.0 |
50.0 |
0.5 |
|
20.25 |
80.0 |
20.0 |
0.25 |
|
25.25 |
80.0 |
20.0 |
0.25 |
|
27.25 |
22.0 |
78.0 |
0.5 |
|
31.00 |
22.0 |
78.0 |
0.5 |
|
Mass spectrometer: |
Xevo G2 QTof |
|
Ionization mode: |
ESI+ |
|
Analyzer mode: |
Sensitivity |
|
Capillary voltage: |
3.20 kV |
|
Cone voltage: |
37 V |
|
Source temp.: |
100 °C |
|
Desolvation temp.: |
350 °C |
|
Cone gas flow: |
0.0 L/h |
|
Desolvation gas flow: |
800 L/h |
|
Calibration: |
NaI, 1 μg/μL from 50 to 2000 m/z |
|
Acquisition: |
700 to 3000 m/z, 1 Hz scan rate |
|
Lock mass: |
0.5 μM [Glu1]-fibrinopeptide in 50:50 ACN/water, 0.1% formic acid |
UNIFI and MassLynx software
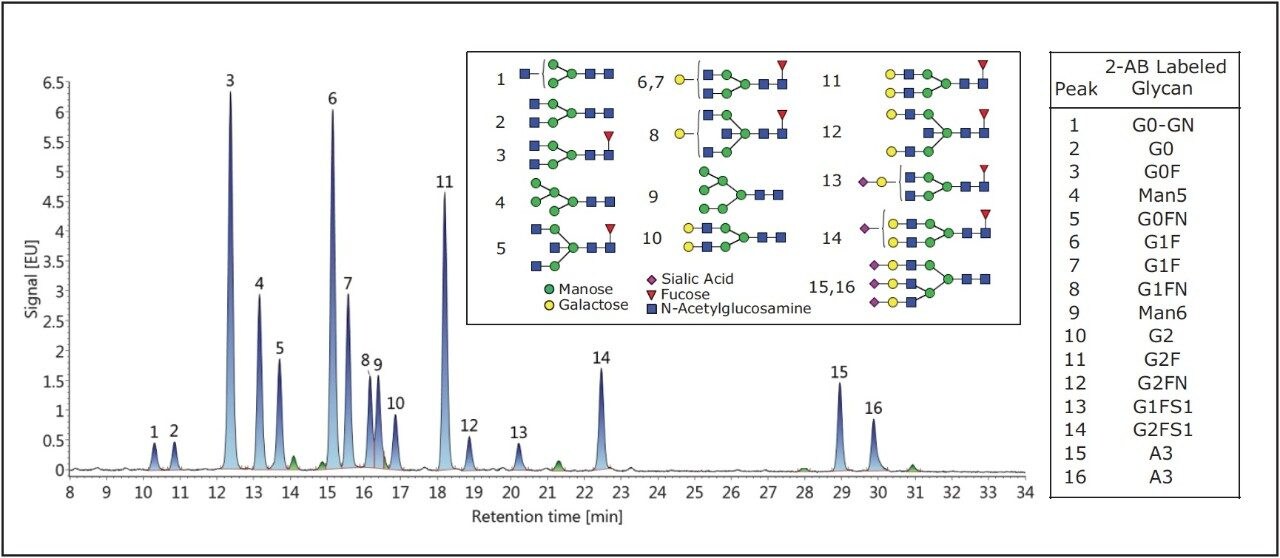
A test mixture, capable of rigorously interrogating the recovery of N-glycans from GlycoWorks HILIC SPE, was prepared by combining the Glycan Performance Test Standard with 2-AB labeled trisialylated A3 glycans. The Glycan Performance Test Standard is comprised of 2-AB labeled N-glycans derived from pooled human serum IgG spiked with high mannose glycans (Man5 and Man6). The addition of the trisialylated A3 glycans further extends the complexity of this mixture, as the A3 glycans are larger, more acidic, and bind more strongly in a HILIC-based separation than glycans commonly found on human or human-like IgG. Figure 2 shows a HILIC-FLR analysis of this modified test mixture using an ACQUITY UPLC GST Amide (BEH Glycan) Column along with UNIFI Software for instrument control and data interpretation.
Based on this analytical approach, the HILIC SPE of the GlycoWorks solution was evaluated. A silica-based aminopropyl sorbent is contained in the GlycoWorks Kit (p/n 176003090). This sorbent was selected from several tested because it is highly polar and, consequently, useful for HILIC separations. Since this sorbent possesses a weakly basic surface and potential for anion exchange, it was, however, assumed that the relative and total recovery of glycans from a GlycoWorks HILIC SPE device could be particularly sensitive to elution conditions. To evaluate this step, elution from the GlycoWorks HILIC sorbent was studied in detail. 2-AB labeled glycans were loaded onto a 96-well HILIC μElution Plate according to the protocol provided in the GlycoWorks High-throughput Sample Preparation Kit Care and Use Manual.13 Various eluents were then employed for elution of the labeled glycans, and recoveries for each major species in the test mixture was subsequently determined. These data were compared alongside the recoveries of the glycans from just the lyophilization and reconstitution steps that were performed after the HILIC SPE procedure, in preparation of the samples for HILIC-FLR. A series of eluents comprised of 20% ACN and increasing concentrations of ammonium bicarbonate (NH4HCO3, pH 8–9) were first investigated. A volatile salt was chosen, due to requisite lyophilization steps. Interpretation of the recoveries led to the observation that the recovery of the glycans was biased, based on eluent choice, with smaller, neutral species recovered better than larger, acidic species. With an eluent comprised of simply 20% ACN/80% water (H20) and no other components, acidic glycans in the test mixture were either poorly recovered or not recovered at all; meanwhile, neutral glycans were obtained with reasonable recovery (≥70%). The addition of NH4HCO3, to concentrations of 25 mM or higher minimized this apparent and non-desired ionic retention mechanism. Nevertheless, even with 100 mM NH4HCO3, there was a noticeable correlation between recovery and the hydrophilicity, or glucose unit (GU) values, of the glycans (Figure 3A).
Biased recovery, or speciation, can be problematic for a sample preparation procedure. In addition to not providing an accurate representation of the species present in the sample, it can be indicative of a method that is not robust and that the relative abundance profiles obtained may not be reproducibly determined, particularly with respect to the most poorly recovered species. As a result, a study was performed to improve these observed 2-AB labeled glycan recoveries. Given that retention of polar analytes to a polar sorbent is dominated by hydrogen bonding and ionic interactions, eluents with more aqueous content (decreased ACN concentrations) were evaluated (Figure 3B). As predicted, NH4HCO3 eluents comprised of lower concentrations of organic solvent yielded both higher and less biased recoveries of the glycan profile. Within the range of this study, an eluent composition of 25 mM NH4HCO3 /5% ACN was found to produce optimal recoveries.
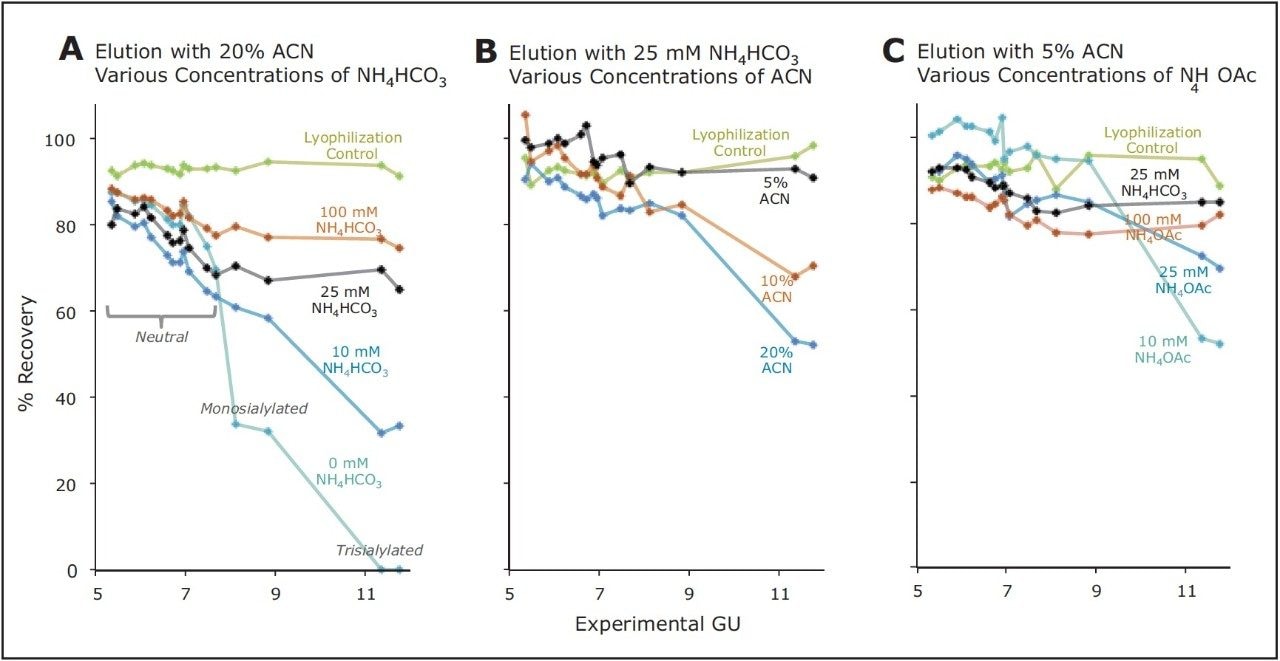
Unfortunately, eluents containing NH4HCO3 posed a challenge in this application as their basicity (typically pH 8 but increased toward pH 9 upon exposure to air) may result in noticeable dissolution of the silica SPE particles and problematic levels of precipitate in the reconstituted samples. To eliminate this potential issue and establish a more robust procedure, we investigated alternative eluents based on neutral solutions of ammonium acetate (pH 7). The effect of ammonium acetate (NH4OAc) eluents on the recoveries of the 2-AB labeled glycans is shown in Figure 3. A 100-mM NH4OAc, 5% ACN eluent was selected as the optimal elution condition, since it provided high as well as relatively unbiased analyte recoveries, similar to those obtained using the 25-mM NH4HCO3, 5% ACN eluent.
The set of chromatograms shown in Figures 4A and 4B demonstrates that the test mixture, before and after HILIC SPE treatment, exhibits highly consistent glycan profiles. Relative abundance determinations for control samples as well as a processed sample are shown in Figure 4C. Compared to the control, differences in relative abundances were ≤7% across the entire profile. For example, the relative abundance of G0F (peak 3) was determined to be 17.9% and 18.6%, before and after SPE, respectively. The relative abundance of trisialylated A3 (peak 16) before and after SPE was 2.8% and 2.7%, respectively (Figure 4C). These optimized elution conditions provide quantitative recoveries for both glycans typical of human IgGs and heavily sialylated glycans, as demonstrated with the recovery of the A3 glycan. With these conditions, the GlycoWorks HILIC μElution Plate is well suited for the preparation of N-glycans from a range of glycoproteins, including those with primarily low GU value neutral glycans as well as those decorated with high GU value, heavily sialylated glycans.
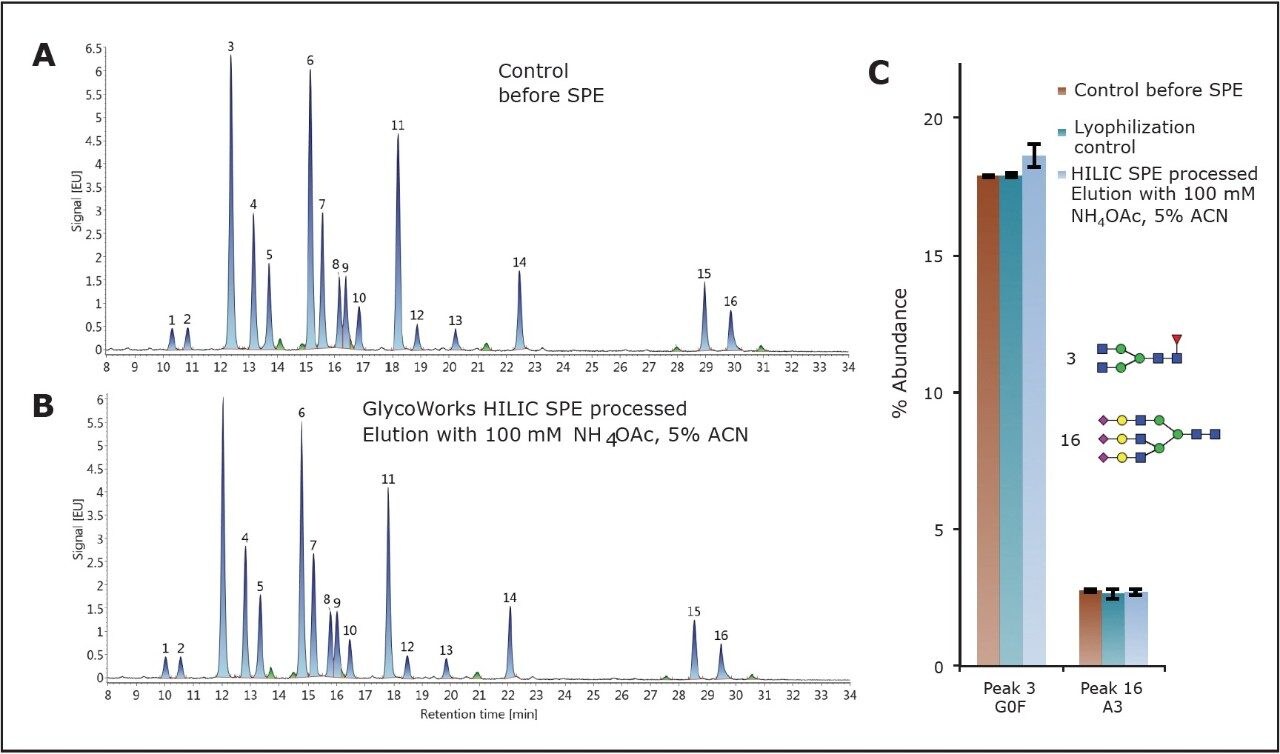
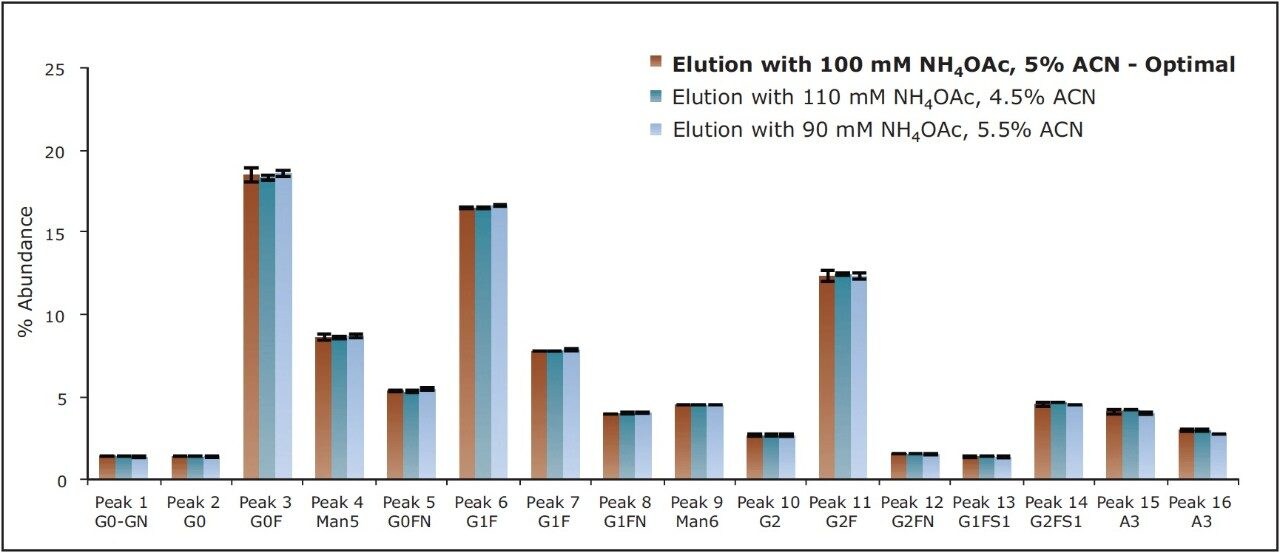
The GlycoWorks HILIC μElution Plate was optimized to yield desired recoveries and, more importantly, to be robust. Elution conditions were purposely optimized so that even relatively large changes in critical elution parameters, namely organic concentration and ionic strength, would have minimal effect on the obtained glycan profile. To demonstrate this, the HILIC SPE method was subjected to robustness testing. Glycan profiles obtained using SPE eluents with the optimized concentrations of ACN and NH4OAc concentrations were compared to those obtained with eluents comprised of ACN and NH4OAc concentrations varied by 10%. The impact of changes in ionic strength and ACN concentration were purposely compounded in these studies. A strong eluent with 110-mM NH4OAc, 4.5% ACN as well as a comparatively weak eluent with 90-mM NH4OAc, 5.5% ACN were employed. Figure 5 shows the relative abundances for each of the major constituents in the test mixture obtained using these varied conditions. The glycan profiles obtained were comparable to the conditions tested. The largest percent change observed between relative abundances from the optimal to extreme conditions was only 7%, corresponding to the recovery of trisialylated A3 (peak 16). This result demonstrates that clean-up of 2-AB labeled glycans using the GlycoWorks HILIC μElution Plate with the optimized elution conditions exhibits noteworthy ruggedness, and is, therefore a robust solution for N-glycan preparations even in quality control applications.
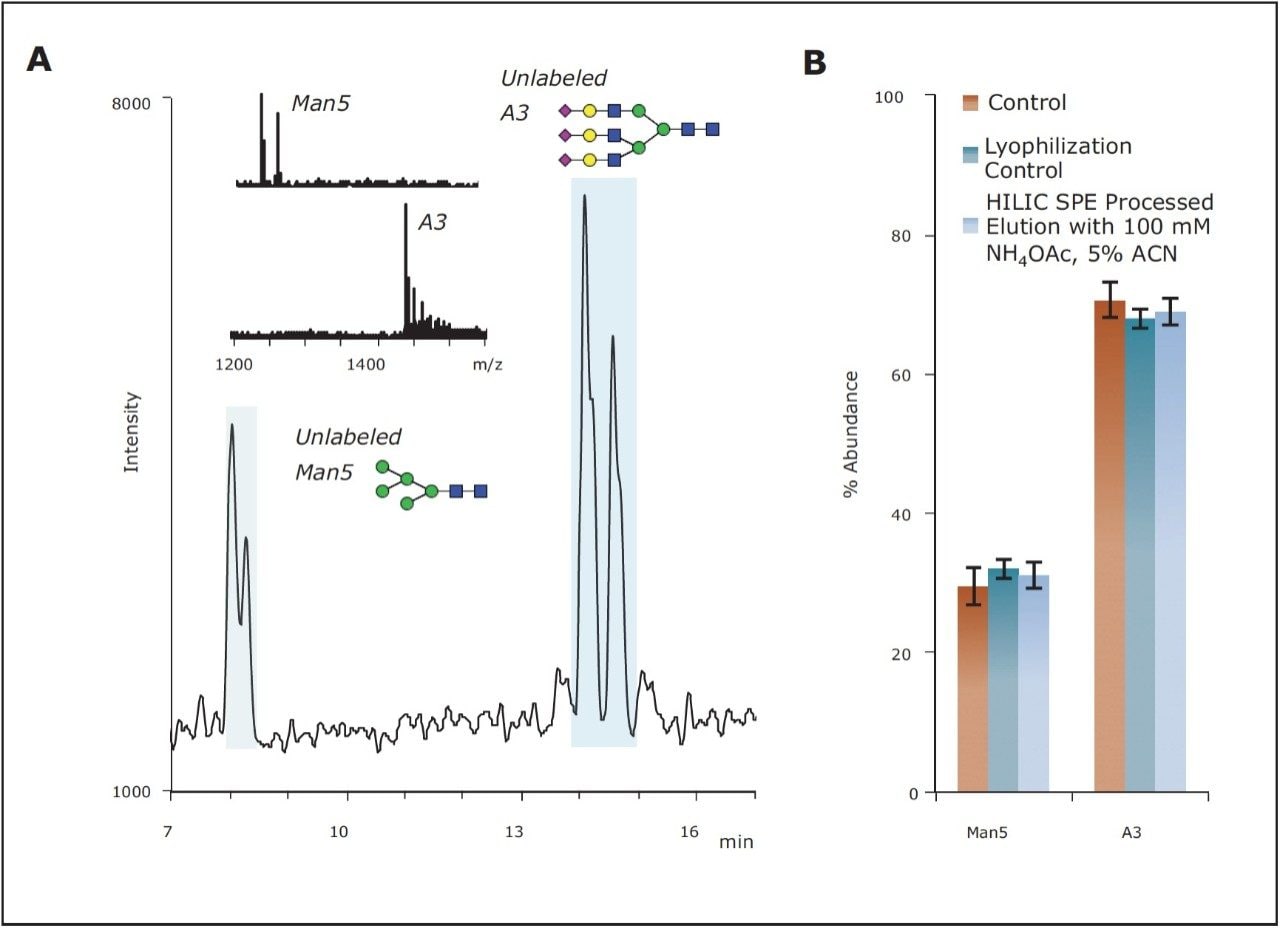
The GlycoWorks HILIC μElution Plate is also suggested for the initial purification of unlabeled glycans cleaved from the target glycoprotein via enzymatic digestion. 2-AB labeled glycans, as previously studied, are slightly less hydrophilic than unlabeled glycans due to the hydrophobicity of the benzamide fluorescent tag. To confirm that recoveries of unlabeled glycans were similar in comparison to the recoveries of 2-AB labeled glycans, an additional study was performed. A HILIC-MS assay was established to determine the relative abundances of two unlabeled glycans representing the extremes of most IgG N-glycan profiles. The mixture tested was comprised of equal amounts of a neutral, low GU value glycan (Man5) and an acidic, high GU value glycan (trisialylated A3). An extracted ion chromatogram (XIC) obtained for this mixture with a Xevo G2 QTof is shown in Figure 6A. Interestingly, two major peaks were observed for both unlabeled Man5 and A3, indicating the presence of different isoforms. Mass spectral windows wide enough to capture both protonated and salt adductspecies of the unlabeled glycans were used to construct the chromatogram. XICs obtained in this manner were integrated, and the obtained peak areas were used to calculate relative abundances of the unlabeled glycans before and after HILIC SPE (Figure 6B). As with 2-AB labeled glycans, the profile of the unlabeled glycanmixture before and after SPE was highly comparable, indicating that the optimized GlycoWorks HILIC SPE process also yields minimally biased recoveries of unlabeled glycans.
HILIC SPE was rigorously studied and optimized to provide quantitative recoveries of 2-AB labeled and unlabeled N-glycans. A test mixture containing a diverse array of 2-AB labeled N-glycans was employed to interrogate GlycoWorks HILIC μElution Plate performance, and develop optimized elution conditions for a robust and reproducible method. In ruggedness testing of the optimized SPE, only minimal changes in a glycan profile were observed despite significant changes in the critical parameters of the SPE eluent. Moreover, an LC-MS assay showed that unlabeled glycans, like 2-AB labeled glycans, are recovered with minimal bias using the newly optimized elution conditions. These studies highlight the development of the GlycoWorks solution and its value in facilitating the release, labeling, and purification of N-glycans.
720004717, June 2013