For research use only. Not for use in diagnostic procedures.
This application note describes a qualitative and quantitative protein identifications using SONAR mode of acquisition.
A comparative study of DIA methodologies show qualitative improvements with increased protein identification numbers for data acquired using SONAR when compared against SWATH. Results based on varying gradient length showed SONAR to provide significantly more protein identifications than SWATH, particularly when adopting a shorter chromatographic timescale, i.e. the utilization of capillary LC for improved throughput and robustness. Reducing the gradient from 60 to 30 min resulted in only a marginal difference in protein identification rates for SONAR, whereas comparative SWATH data showed a decrease of 23%. The rapidly scanning nature of the quadrupole for SONAR also provides quantitative benefits, with precision being maintained for high throughput analysis.
Qualitative improvements with increased protein identification numbers for data acquired using SONAR when compared against SWATH. SONAR provides significantly more protein identifications than SWATH, particularly when adopting a shorter chromatographic timescale, i.e. the utilization of capillary LC for improved throughput and robustness.
The continued requirement to analyze larger sample cohorts to detect quantitative biologically significant differences is becoming of greater importance and placing greater demands on instrument time. Traditionally, proteomic LC-MS analyses have been conducted using nanoscale chromatography in combination with data dependent analysis (DDA). However, the adoption of faster chromatography to increase sample throughput and data independent approaches (DIA) are proving increasingly popular. A number of DIA strategies with enhanced specificity exist, such as SWATH, whereby the quadrupole is stepped across a mass range of interest to increase specificity. However, this approach can have drawbacks when utilizing faster chromatographic methods since the duty cycle of the instrument is challenged. An alternate DIA method, which also uses a quadrupole analyzer for additional selectivity, is SONAR, whereby the quadrupole is scanned as opposed to being stepped over the mass range of interest. The fast scanning nature of the method makes the technique particularly suited for fast chromatographic, high throughput workflows. Here, we present results from a comparative DIA experiment set using a tryptic digest of the K562 cell line, separated using capillary scale chromatography and MS data acquired using SWATH (stepped quadrupole) and SONAR (scanning quadrupole) modes of acquisition.
K562 cell line tryptic digest standard (Sigma-Aldrich, St. Louis, MI)
|
LC system: |
M-Class ACQUITY UPLC |
|
Column(s): |
1.8 μm HSS T3 C18 300 μm x 100 mm NanoEase analytical |
|
Column temperature: |
35 °C |
|
Flow rate: |
7 μL/min |
|
Mobile phase: |
Water (0.1% formic acid) (A) and acetonitrile (0.1% formic acid) (B) |
|
Gradient: |
3% to 40% B in 30 or 60 min |
|
Injection volume: |
1 μL (1, 5 or 10 μg) |
|
MS system: |
Xevo G2-XS QTof |
|
Ionization mode: |
ESI (+) at 3.2 kV |
|
Cone voltage: |
30 V |
|
Acquisition mode: |
SONAR |
|
Acquisition range: |
50 to 2000 m/z both functions (low and elevated energy) |
|
Acquisition rate: |
0.5 s both functions (low and elevated energy) |
|
Quadrupole scan range: |
400 to 900 m/z |
|
Isolation window: |
24 Da |
|
Collision energy: |
5 eV (low energy function) and from 19 eV to 45 eV (elevated energy function) |
|
Resolution: |
35,000 FWHM |
|
MS system: |
Sciex TripleTOF 5600 |
|
Acquisition mode: |
SWATH 50 m/z to 2000 m/z both functions |
|
Quadrupole window: |
60 target windows (variable) stepping quadrupole with 1 Da overlap |
The LC-MS data were processed with Mascot Distiller (Matrix Science, London, United Kingdom) and Spectronaut Pulsar (Biognosys AG, Schlieren, Switzerland). Data were searched against a study specific K562 library.
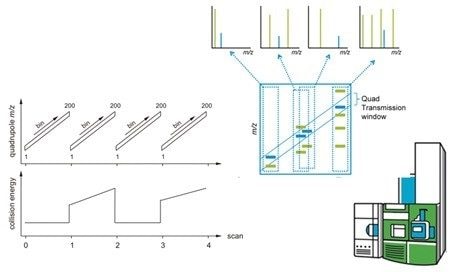
The principle of SONAR, a scanning quadrupole based data independent acquisition (DIA) method, is illustrated in the left hand side image of Figure 1. In short, alternate datasets are acquired in low (MS1) and elevated (MS2) collision energy mode.1,2 During each low and elevated energy segment, the quadrupole isolation window is scanned linearly between two user-selected positions and 200 TOF spectra are acquired. The quadrupole scan duration is application/chromatographic peak width dependent and typically varies from 0.1 s to 1 s. In the elevated energy mode, the collision energy can be ramped between two values, which are selected to optimize fragmentation efficiency at each quadrupole position. The selectivity of the acquisition method is illustrated by the middle image where, dependent on the position of the quadrupole, i.e. transmission m/z window, precursor (even those close in mass) and product ions can be exclusively isolated.
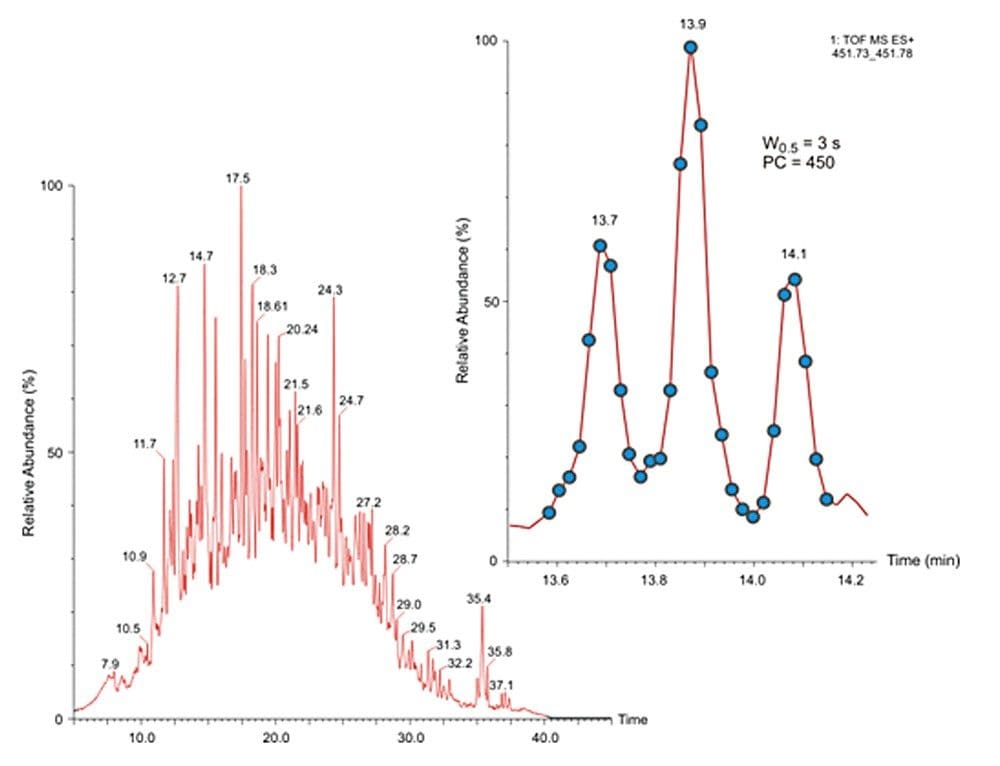
The requirement for acquisition speed is clearly demonstrated in Figure 2, showing a 50 Da wide mass extracted chromatogram for a 30 min reversed phase gradient separation of non-fractionated K562 tryptic digest. Shown inset is a 10 mDa wide mass extracted chromatogram over a narrow chromatographic window of 30 s. Typical peak widths at half height were 3 s; hence, to retain a sufficient number of points across the peaks for precise quantitation while maintaining optimum S/N, the scan speed was set to 0.5 s, providing between six and eight data points across a peak. The peak capacity for 30 min high throughput proteomic separations was estimated to be ~ 450. The importance of peak sampling frequency and its effect on quantitative precision is described in more detail elsewhere.1
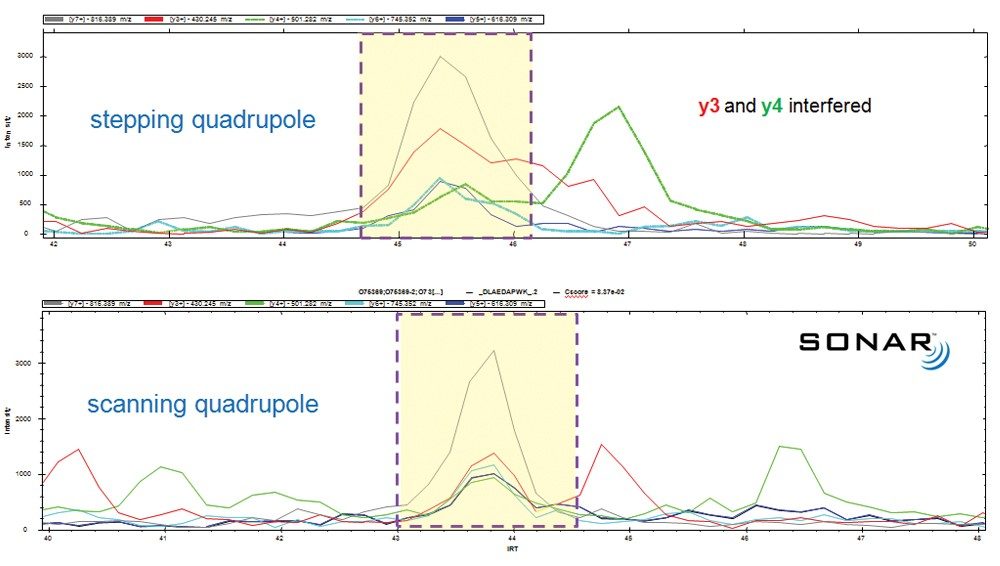
The benefit of high speed quadrupole scanning vs. quadrupole stepping with respect to selectivity is presented in Figure 3, where the same sample and amount were analyzed under identical chromatographic conditions. Five mass extracted product ion chromatograms corresponding to fragment ions of the same peptide are shown. The upper pane, representing a stepped quadrupole SWATH acquisition, identified all five product ions of interest; however, it can also be seen that both y3 and y4 were interfered, affecting quantitative precision. In contrast, the SONAR method afforded non-interfered detection and extraction of the chromatographic of the same set of fragment ions.
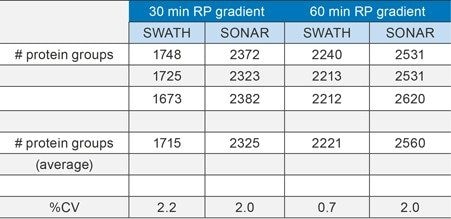
Increased acquisition speed also has a profound effect on qualitative performance, especially in the instance of limited amount(s) and/or increased throughput experiments. The results shown in Figure 4 and Table 1 summarize the number of K562 protein groups (based on a 1 µg loading) identified by Spectronaut informatics. Interpretation of 30 and 60 min gradient separations for three technical replicates indicates a significant increase in protein identifications for SONAR when comparing the two techniques. For the 60 min gradient, approximately 300 additional proteins are identified with SONAR; however, greater gains are observed with the shorter 30 min gradient, with approximately 600 more identifications being achieved when implementing SONAR. The confidence of the identifications in both cases is further exemplified with the peptide retention alignment precision between the observed and library entries.
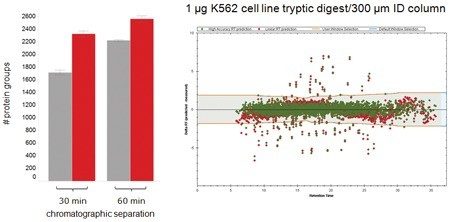
A comparative study of DIA methodologies show qualitative improvements with increased protein identification numbers for data acquired using SONAR when compared against SWATH. Results based on varying gradient length showed SONAR to provide significantly more protein identifications than SWATH, particularly when adopting a shorter chromatographic timescale, i.e. the utilization of capillary LC for improved throughput and robustness. Reducing the gradient from 60 to 30 min resulted in only a marginal difference in protein identification rates for SONAR, whereas comparative SWATH data showed a decrease of 23%. The rapidly scanning nature of the quadrupole for SONAR also provides quantitative benefits, with precision being maintained for high throughput analysis.
720006161, January 2018