For research use only. Not for use in diagnostic procedures.
This is an Application Brief and does not contain a detailed Experimental section.
This application note demonstrates the improved spectral clarity achieved when using SONAR and thereby enabling increased identification accuracy of co-eluting compounds for complex lipidomic studies.
Implementing SONAR improves spectral clarity and thereby increases certainty of lipid identifications.
Lipidomic analysis of blood and tissue samples for biomedical research aims to identify and quantify potential biomarkers or analytes of interest. When investigating these biological samples via LC-MS, high collision energy fragmentation data is essential for the identification of significant features using database searches. Current data independent acquisition (DIA) modes switch between low and high collision energy functions in order to generate fragmentation spectra. Due to the complex nature of biological matrices, liquid chromatography (LC) is employed to better resolve the components prior to analysis by mass spectrometry (MS). However, due to the complexity of the lipidome, many lipids co-elute as a result of chemical similarities within lipid classes, and therefore undergo simultaneous MS/MS fragmentation. This results in high energy spectra comprised of fragments from multiple precursor ions, making it difficult to attribute which fragments correspond to which precursor ions, and therefore increasing the difficulty of making accurate identifications. SONAR is a DIA technique with a low-resolution quadrupole mass filter, which is scanned repetitively, and both precursor and MS/MS data are acquired at spectral rates approaching 2000 spectra per second (Figure 1). Here we describe the application of SONAR for the investigation of chimeric humanized mouse liver extracts.
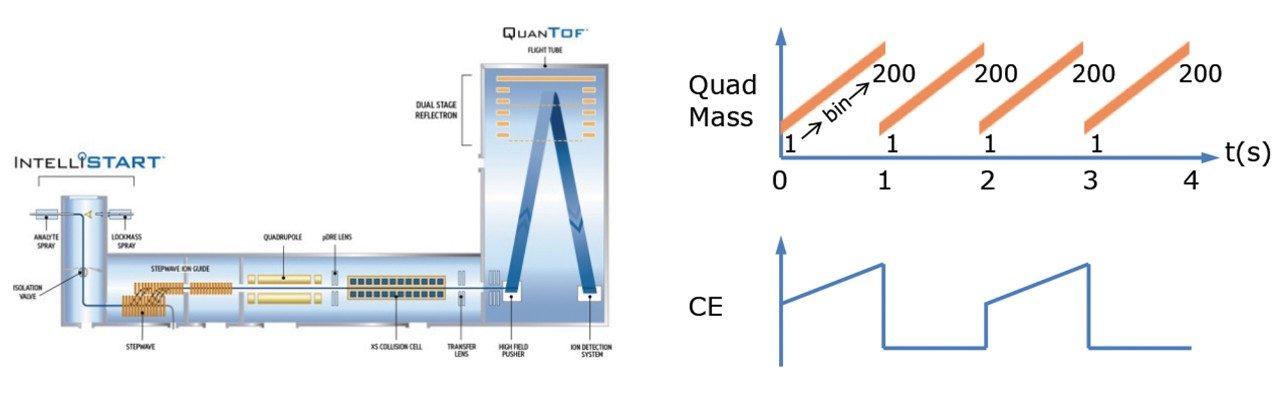
SONAR has been utilized for the analysis of lipid extracts derived from chimeric humanized mouse livers. Lipids were extracted from 17 livers using Dichloromethane (DCM):Methanol (3:1) and subsequently evaporated using a rotary evaporator. Once dry, the extracts were reconstituted in Isopropyl Alcohol (IPA):Methanol:Water prior to analysis. A pooled sample was prepared from each extract and used as a quality control (QC) which was injected every seventh injection. Data were also collected using an alternative DIA method for comparative purposes. Lipids were separated over a 20 minute LC gradient using reversed phase (RP) chromatography (2.1 x 100 mm ACQUITY UPLC CSH C18 Column, 1.7 µm). LC-MS data were acquired using a Xevo G2-XS QTof mass spectrometer operating in SONAR mode with a quadrupole window of 10 Da, scanning over a mass range of 350 – 950 m/z, while the Tof scanned over 50 – 1200 m/z with a scan rate of 0.1 sec.
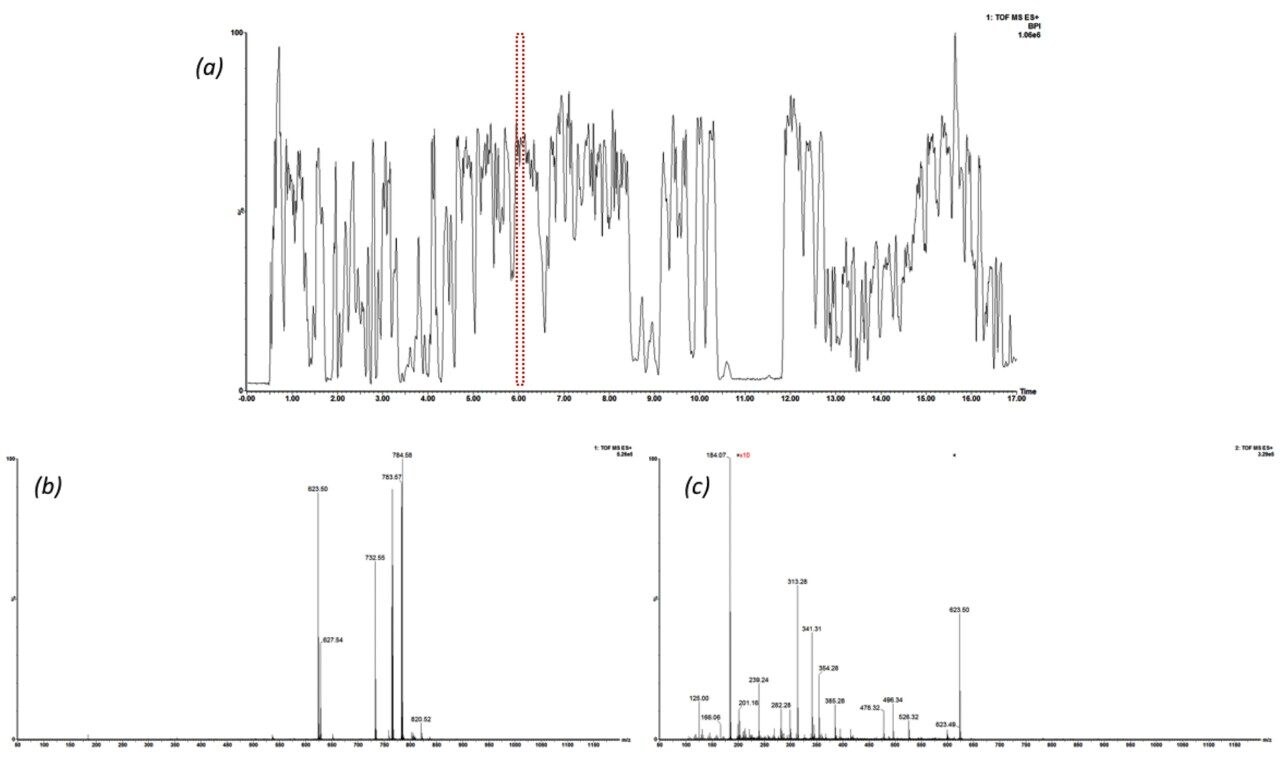
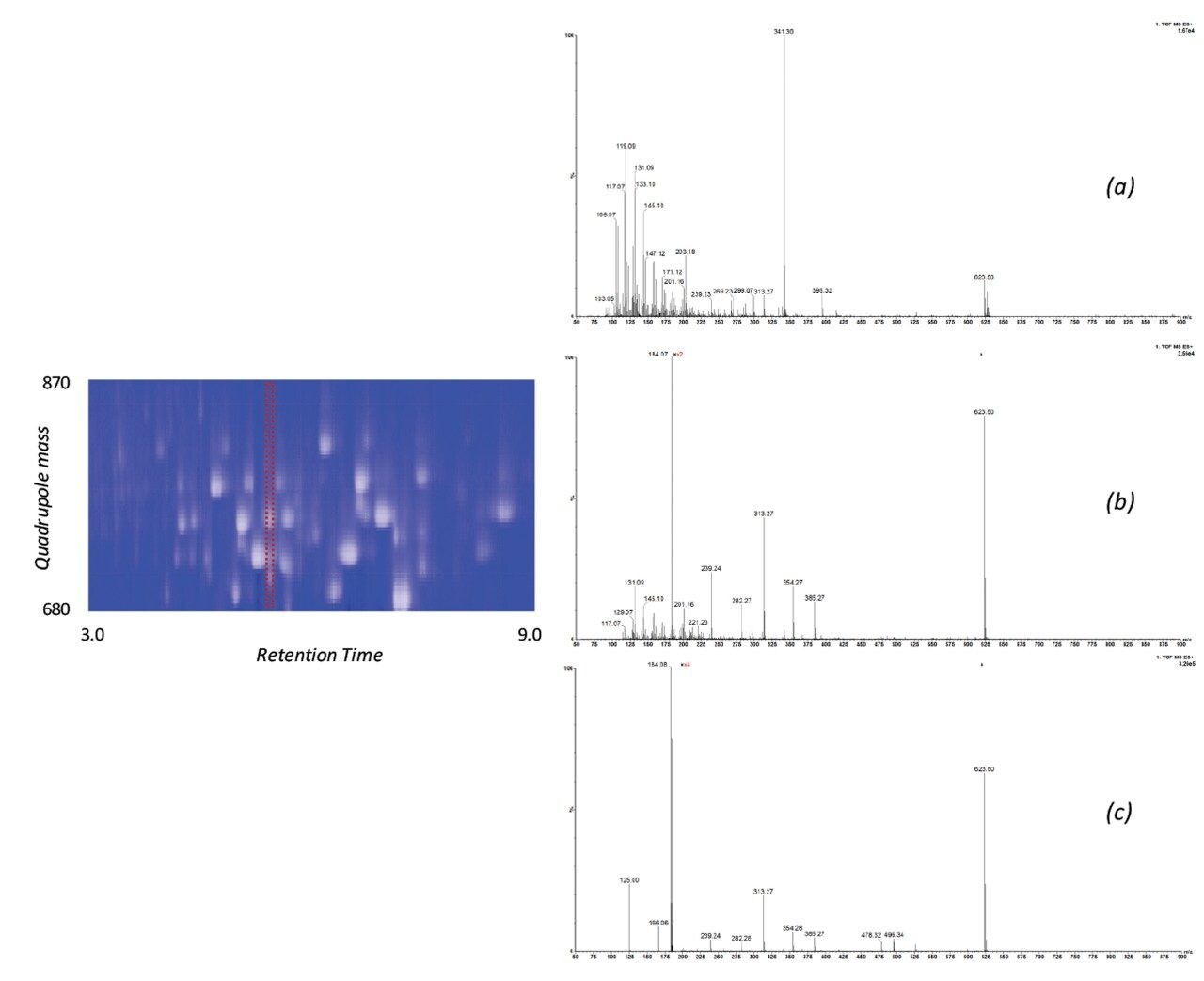
The issue of co-elution is evident with multiple precursors generating mixed fragmentation spectra. The data displayed in Figure 3, however, represents SONAR generated data highlighting the specificity of the technique by resolving co-eluting lipid species. The acquisition mode utilizes a scanning quadrupole, filtering ions over a set m/z range, providing significantly cleaner spectra when compared to conventional DIA methods. Representative MS and MS/MS spectra of three co-eluting lipids show the additional specificity afforded by the scanning quadrupole technique when compared with an alternative DIA method, which uses a non-resolving quadrupole. Data from all experiments were subsequently peak picked and database searched using Progenesis QI. For regions of high co-elution, a large number of false positive identifications can result from compound database searches. However, the high selectivity provided by SONAR shows the number of false positives to be significantly reduced while increasing the confidence score of the identifications returned when compared with conventional DIA methods. Figure 4 illustrates this finding, showing an increased number of lipid identifications with higher scoring.
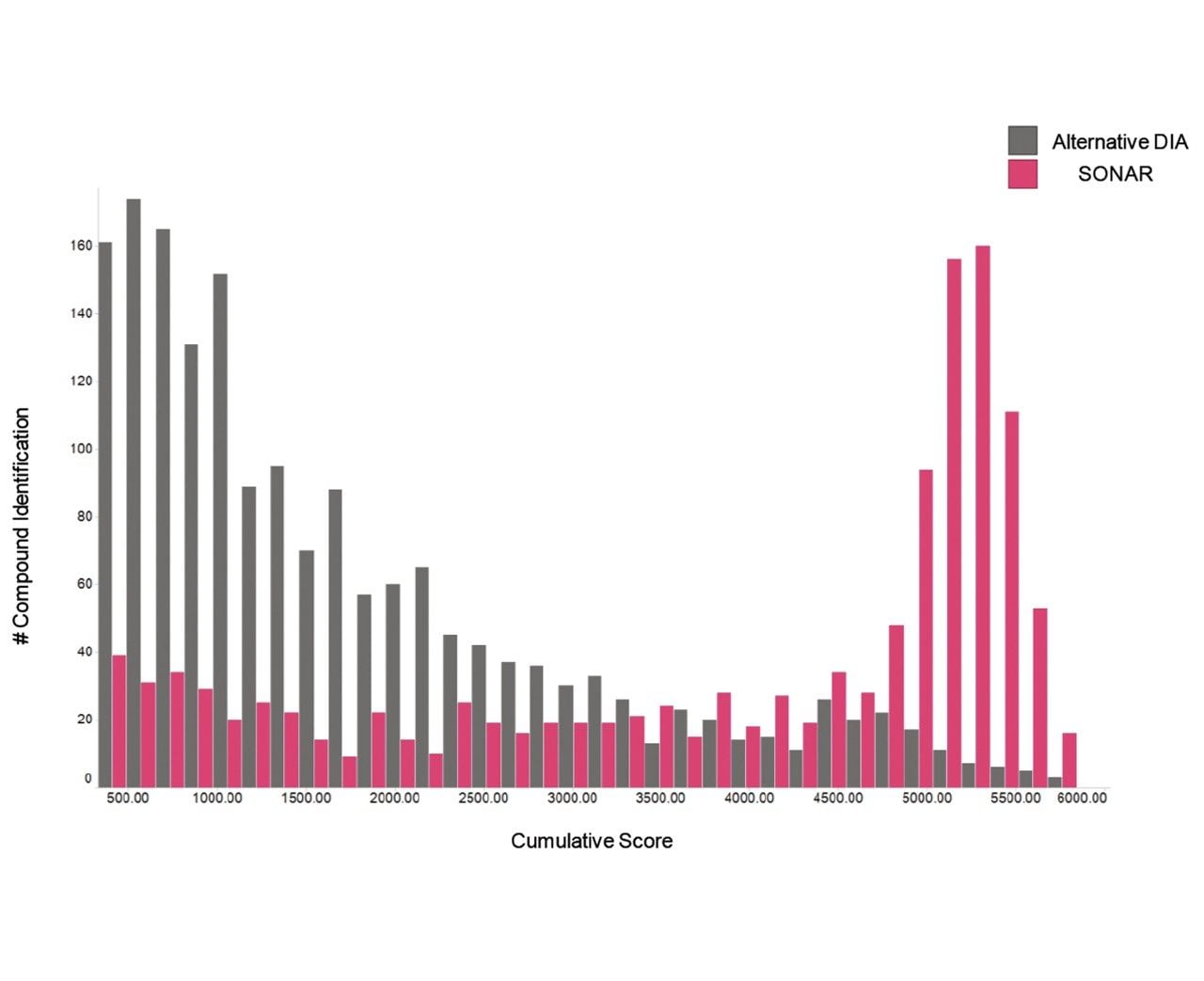
This study demonstrates the benefits of SONAR for omics studies, providing enhanced specificity, clearer spectra, and ultimately fewer false positives when conducting compound database searches. A comparison of SONAR with an alternative DIA methodology shows how increased specificity in the workflow can reduce false positives that result from simultaneous fragmentation from multiple species within the collision cell.
720006036, June 2017