This application note describes an easy to use method that employs mass detection coupled with liquid chromatography for screening chloramphenicol in milk.
Chloramphenicol is an inexpensive broad spectrum antibiotic. In certain susceptible individuals, chloramphenicol has been associated with toxic effects. It is suspected to be both a carcinogen and genotoxic compound, and cases of fatal aplastic anemia (depression of bone marrow) are widely documented. Therefore, its use in food producing animals, insects, and aquaculture is prohibited in the EU, Canada, the U.S., along with a number of Asian and South American countries. Within the EU, Commission Decision 2003/181/EC requires that any method for the detection of chloramphenicol in food have a minimum required performance limit (MRPL) of 0.3 μg.kg-1.
Dairy testing laboratories traditionally screen large volume samples using enzyme-linked immunosorbent assay (ELISA), where a quick turnaround time is essential given the short shelf life of the product. Although ELISA allows for sensitive and rapid screening of chloramphenicol, one drawback is the significant level of false positive results, where a false positive level as high as 16% has been reported in certain instances.1
Due to cross reaction of the immunosorbent with interfering matrix, chloramphenicol can often be misdetected in milk resulting in the unnecessary destruction of the product. Mass detection offers increased selectivity, which helps differentiate between chloramphenicol and contaminating matrix. Furthermore, mass detection coupled with liquid chromatography (LC) eliminates the need for time-consuming derivatization typically required for screening of chloramphenicol by gas chromatography. The ACQUITY QDa Mass Detector can easily be integrated into LC workflows, with the ease of use that does not require specialist mass spectrometry knowledge.
|
UPLC system: |
ACQUITY UPLC I-Class |
|
Run time: |
5 min |
|
Column: |
ACQUITY UPLC HSS C18 1.8 μm, 2.1 x 100 mm |
|
Mobile phase: |
10 mM ammonium acetate in 45:55 methanol:water |
|
Injection volume: |
10 μL |
|
MS system: |
ACQUITY QDa |
|
Ionization mode: |
ESISIR |
|
channel: |
m/z 321.0 |
|
Cone voltage: |
15 V |
|
Probe temp.: |
Default (600 °C) |
|
Capillary voltage: |
Default (0.8 kV) |
|
Sampling rate: |
Default (5 Hz) |
Semi-skimmed cow’s milk (5 g) samples were weighed into 25-mL centrifuge tubes and fortified to allow for 0.3 μg.kg-1 in the sample. Fat removal and protein precipitation were completed by adding hexane (5 mL) and 70% acetonitrile solution (15 mL) to the sample. Samples were shaken then centrifuged for 10 min at 3,900 G (4 °C). The lower acetonitrile layer (5 mL) was removed and diluted to a total volume of 15 mL with 10 % methanol.
Oasis HLB 3 cc (60 mg) Cartridges were conditioned with methanol (2 mL) and water (4 mL). The diluted samples (15 mL) were loaded on to the cartridge, which was then washed with 5% methanol solution (6 mL). Chloramphenicol was eluted using 100% methanol (6 mL). This eluate was subsequently evaporated under nitrogen, reconstituted with 10% methanol solution (300 μL), and transferred for LC-MS analysis.
Blank milk samples (unfortified) were prepared through the sample method outlined. Once dried under nitrogen, the blank matrix samples were reconstituted using standard solutions of different concentrations to allow for a six-point calibration curve over the range equating to 0.075 to 1 μg.kg-1.
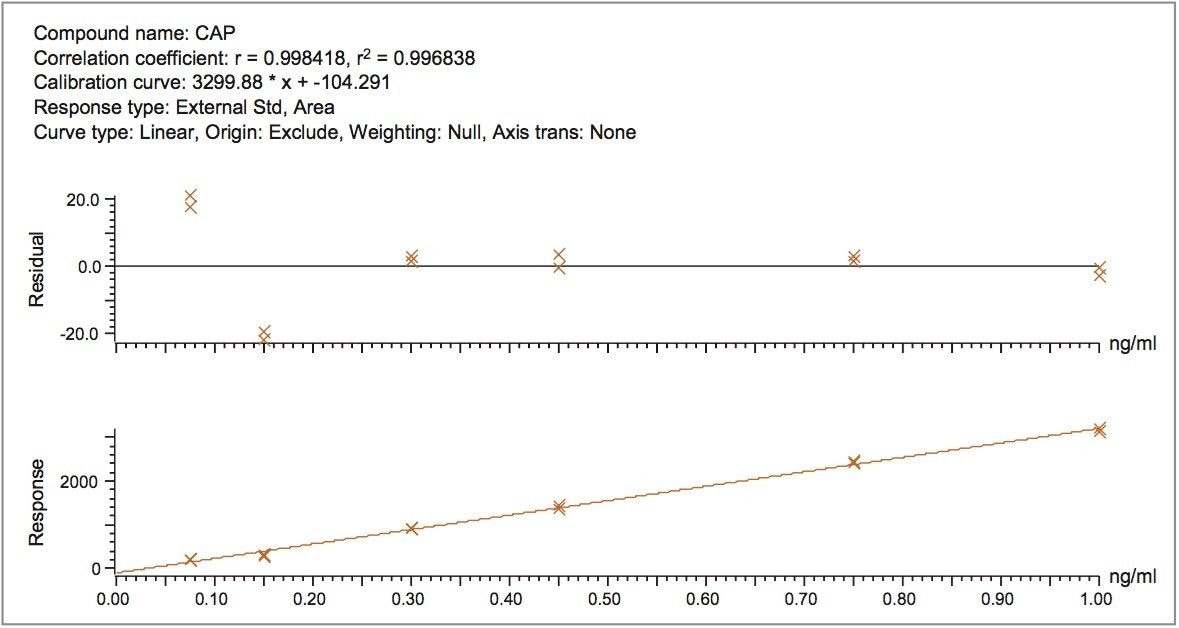
Prior to the analysis of the milk extracts, the optimum source conditions were investigated for chloramphenicol, where the pre-optimized source parameters of the ACQUITY QDa Detector provided for the most efficient and sensitive detection. Chloramphenicol was most sensitive in negative ionization mode with a voltage of 15 applied to the cone. Using the method described, a matrix-matched calibration curve allowed for accurate quantification of milk samples fortified at the EU MRPL. Excellent linearity (R2>0.995) was observed for chloramphenicol over the selected working range of 0.075 to 1 μg.kg,-1 as shown in Figure 1.
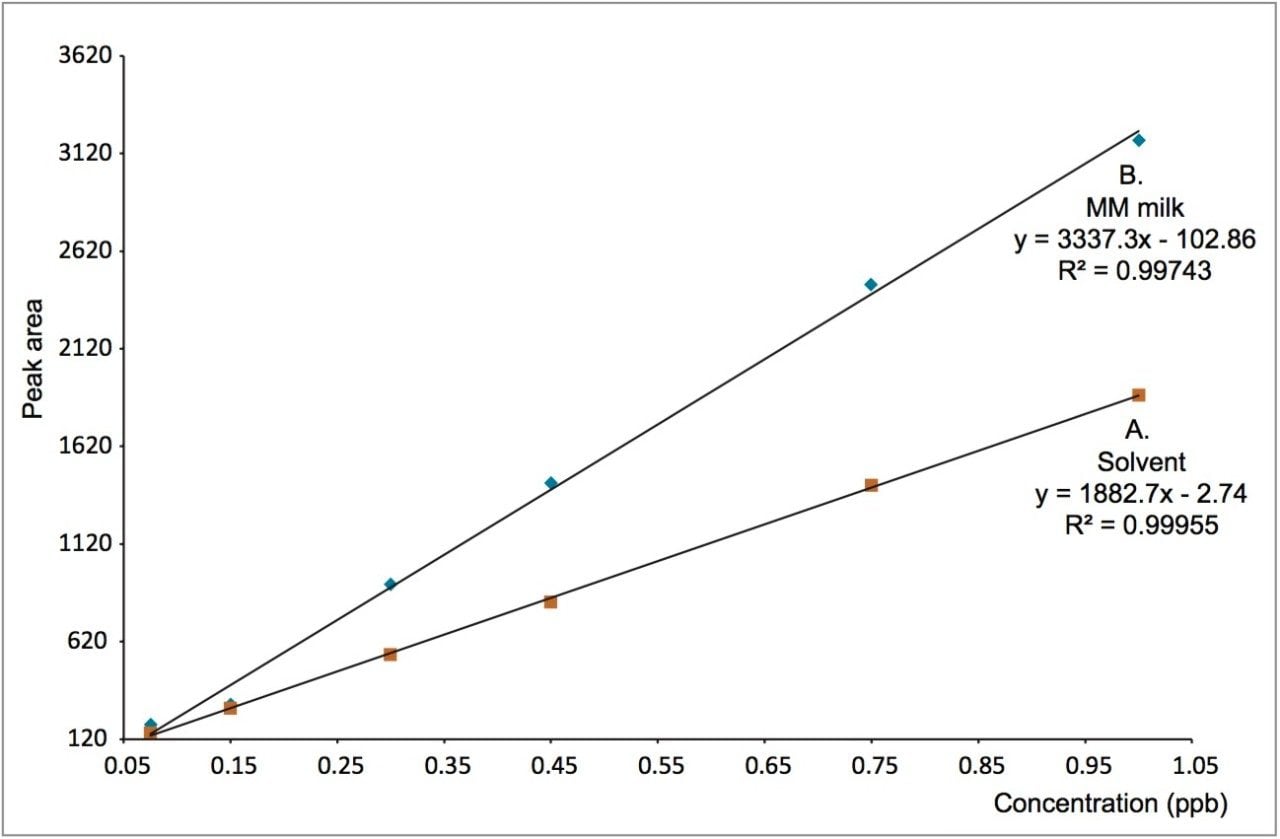
In order to assess matrix effects, the matrix matched calibration curve was compared to a solvent calibration curve over the same range. Figure 2 shows an overlay of the matrix-matched and solvent calibration curves. From this overlay, matrix effects (ion enhancement) are evident. This can also be demonstrated statistically using the slope of each of the calibration curves (slope sample/slope standard x 100), where matrix effects of 177% are calculated, confirming ion enhancement. Despite the matrix effects observed, the accurate quantitative screening of chloramphenicol in milk is readily achievable with the use of the matrix-matched calibration curve.
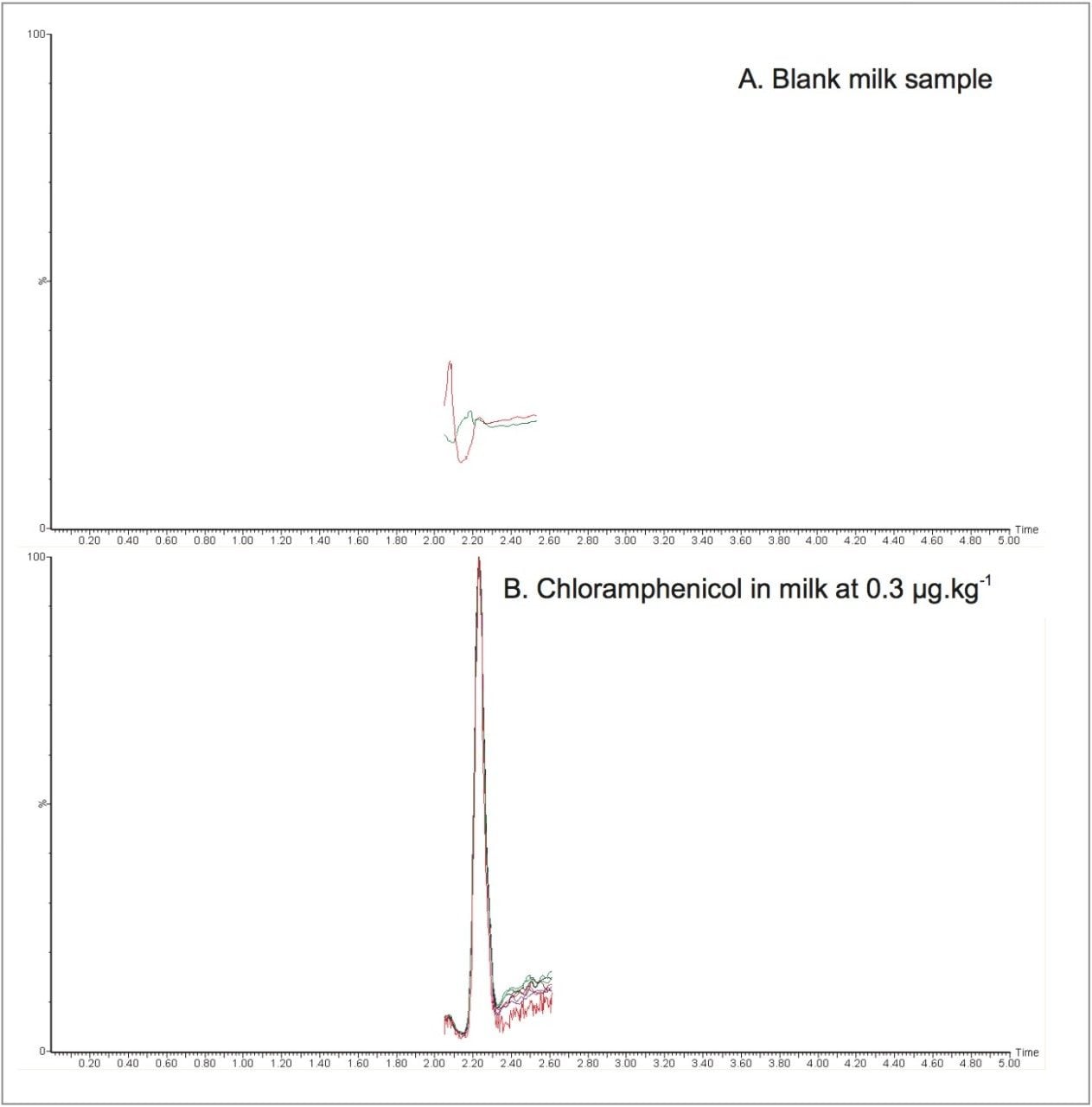
Replicate blank samples of milk were analyzed, where no false positives were detected. An example chromatogram for an extracted blank is shown in Figure 3A, where only baseline noise was observed. Four replicate milk samples were fortified at the EU MRPL, extracted and analyzed in duplicate (n= 8), in the absence of expensive deuterated internal standard. The resultant samples were quantified against the matrix-matched calibration curve. Figure 3B shows the chloramphenicol peak observed for replicates at the fortification level (0.3 μg.kg-1), where good peak-to-peak signal-to-noise ratio (S/N) of ≥50 was achieved.
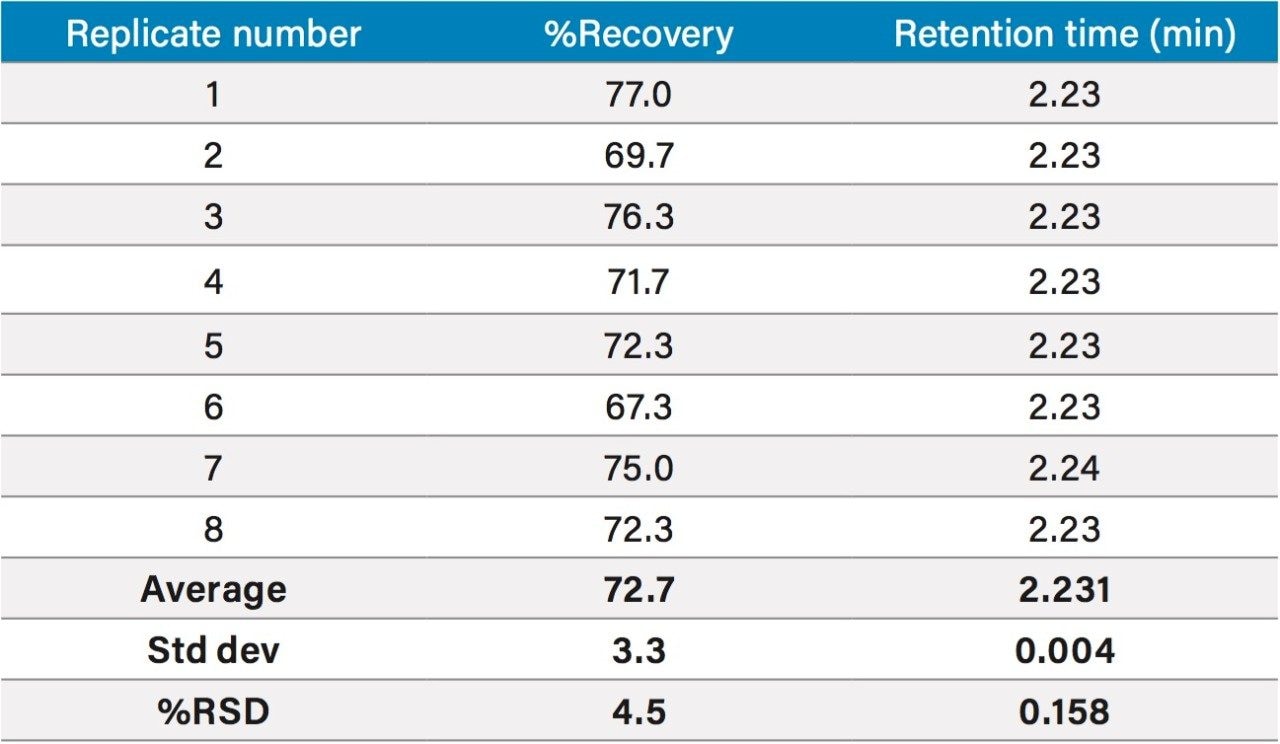
As shown in Figure 3B, good repeatability was observed for the retention time. This is also interpreted statistically, along with recoveries, in Table 1. An average recovery of 72.7% was determined, which is acceptable in accordance with European Commission Decision 2002/657/EC. Excellent repeatability was obtained over eight replicates at the regulatory limit with relative standard deviation (RSD) of <4.6%. This demonstrates that the method is both robust and sensitive for the quantitative screening of chloramphenicol in milk at the regulatory limit.
An accurate and robust method has been developed for the reliable quantitative screening of chloramphenicol in milk. This method has proven to achieve levels of detection that can easily meet regulatory requirements, in the absence of an internal standard, while showing excellent repeatability at the EU MRPL of 0.3 μg.kg-1. The additional discrimination that mass detection affords results in increased specificity, where no false positives were detected.
720004993, November 2016