
In this application note, a multi-residue analysis method for the detection of 212 pesticides in okra is presented.
The combination of ACQUITY UPLC H-Class System with the Xevo TQD tandem mass spectrometer can detect pesticides below the legislative limit in okra samples. Even though a strong matrix effect was observed for many compounds, detection and quantification at the legislative limit was achieved. Simultaneous acquisition of MRMs and RADAR full-scan data provides quantitative and qualitative information in single injection. Product ion confirmation (PICs) increases confidence in compound assignments, which proves highly useful when working with complex matrices.
Okra is an important vegetable of the tropical countries and a popular diet component in several countries including India. According to the Food and Agriculture Organization of the United Nations (FAO),1 India is one of the largest okra producers in the world and it produced approximately 5,800 tons of okra in 2010 and 2011. Okra is susceptible to a variety of pests and diseases2 and a wide-range of pesticides are used to treat okra plants in India. Legislative limits are in place for the presence of pesticides in domestically produced, imported, or exported okra.3 It is, therefore, very important to monitor the presence of commonly used pesticides in okra at legislative limits.
According to the PRiF (Pesticide Residues in Food) report, import controls under regulation (EC) No 669/2009 have been increased for okra imported from India because of the frequent detection of pesticide residues, mainly monocrotophos. The consignment is supposed to be rejected if it is non-compliant with MRLs
(Maximum Residue Limits). Since July 1, 2012, the frequency of testing consignments has been increased from 10% to 50%. With this frequent testing, monocrotophos, triazophos, and acetamiprid were found at 0.02 mg/kg in okra samples from India, while the MRL for these compounds is 0.01 mg/kg.4
In this application note, a multi-residue analysis method for the detection of 212 pesticides in okra is presented. For a complete list of all pesticides, see Appendix A.
A multi-residue MS method for the pesticides was created using Waters Xevo TQD Quanpedia database. All of the pesticides were analyzed under ESI+ or ESI- mode using rapid polarity switching. Full-scan data were acquired in order to assess any matrix effects and the use of two MRMs and product ion confirmation scans were acquired to confirm and quantify the pesticide residues.
|
LC system: |
ACQUITY UPLC H-Class |
|
Column: |
ACQUITY HSS T3, 1.8 μm, 2.1 X 100 mm |
|
Column temp.: |
45 °C |
|
Injection volume: |
10 μL |
|
Flow rate: |
0.45 mL/min |
|
Mobile phase A: |
10 mM ammonium acetate (pH 5) in water |
|
Mobile phase B: |
10 mM ammonium acetate (pH 5) in methanol |
|
Weak needle wash: |
50/50 Water/methanol (v/v) |
|
Strong needle wash: |
10/90 Methanol/water (v/v) |
|
Seal wash: |
90/10 water/methanol |
|
Time (min) |
Flow rate (mL/min) |
%A |
%B |
Curve |
|---|---|---|---|---|
|
Initial |
0.45 |
98 |
2 |
6 |
|
0.25 |
0.45 |
98 |
2 |
6 |
|
12.25 |
0.45 |
1 |
99 |
6 |
|
13 |
0.45 |
1 |
99 |
6 |
|
13.01 |
0.45 |
98 |
2 |
6 |
|
17 |
0.45 |
98 |
2 |
6 |
Table 1. UPLC method for pesticide analysis.
|
MS system: |
Xevo TQD |
|
Ionization mode: |
ESI+/ESI- |
|
Capillary voltage: |
3 kV |
|
Desolvation temp.: |
500 °C |
|
Desolvation gas flow: |
1000 L/Hr |
|
Source temp.: |
150 °C |
Pesticide standards were purchased either from Sigma-Aldrich, Fisher Scientific, or AccuStandard. A mix of all pesticides at 400 ng/mL was prepared in acetonitrile and stored at 4 °C.
QuEChERS is a popular method worldwide for the multi-residue analysis of pesticides in fruits and vegetables. The AOAC official method 2007.01, was used to prepare okra samples that were purchased at a local supermarket. Briefly, okra samples were homogenized in water and 15 grams of homogenate was collected into a 50-mL centrifuge tube. Samples were extracted with acidified acetonitrile and mixed with MgSO4 and NaCl (Tube 1). The tube was shaken for a minute and centrifuged at 1500 rcf for 1 minute. After centrifugation, the matrix cleanup was accomplished by dispersive solid phase extraction (d-SPE) by using 50 mg of primary secondary amine (PSA), 50 mg of C18 bonded silica, 150 mg of MgSO4, and 7.5 mg of graphitized carbon black (GCB).5 1 mL of supernatant from Tube 1 was added to d-SPE cleanup tube and centrifuged at 1500 rcf for 1 minute. 1 mL of this extract was evaporated to dryness and reconstituted in 200 µL of 40/60 acetonitrile/water spiked with internal standard.
All of the pesticides were successfully detected at 10 ppb (0.01 mg/kg) in okra sample. For all of the pesticides, Appendix A lists the ionization mode, retention time, and whether or not the compound was detected in a pre-spike 1 ppb sample, as well as the 10 ppb pre-spike sample. Figure 1 shows an overlay of the total ion chromatogram (TIC) of all the pesticides at 10 ppb in okra sample.

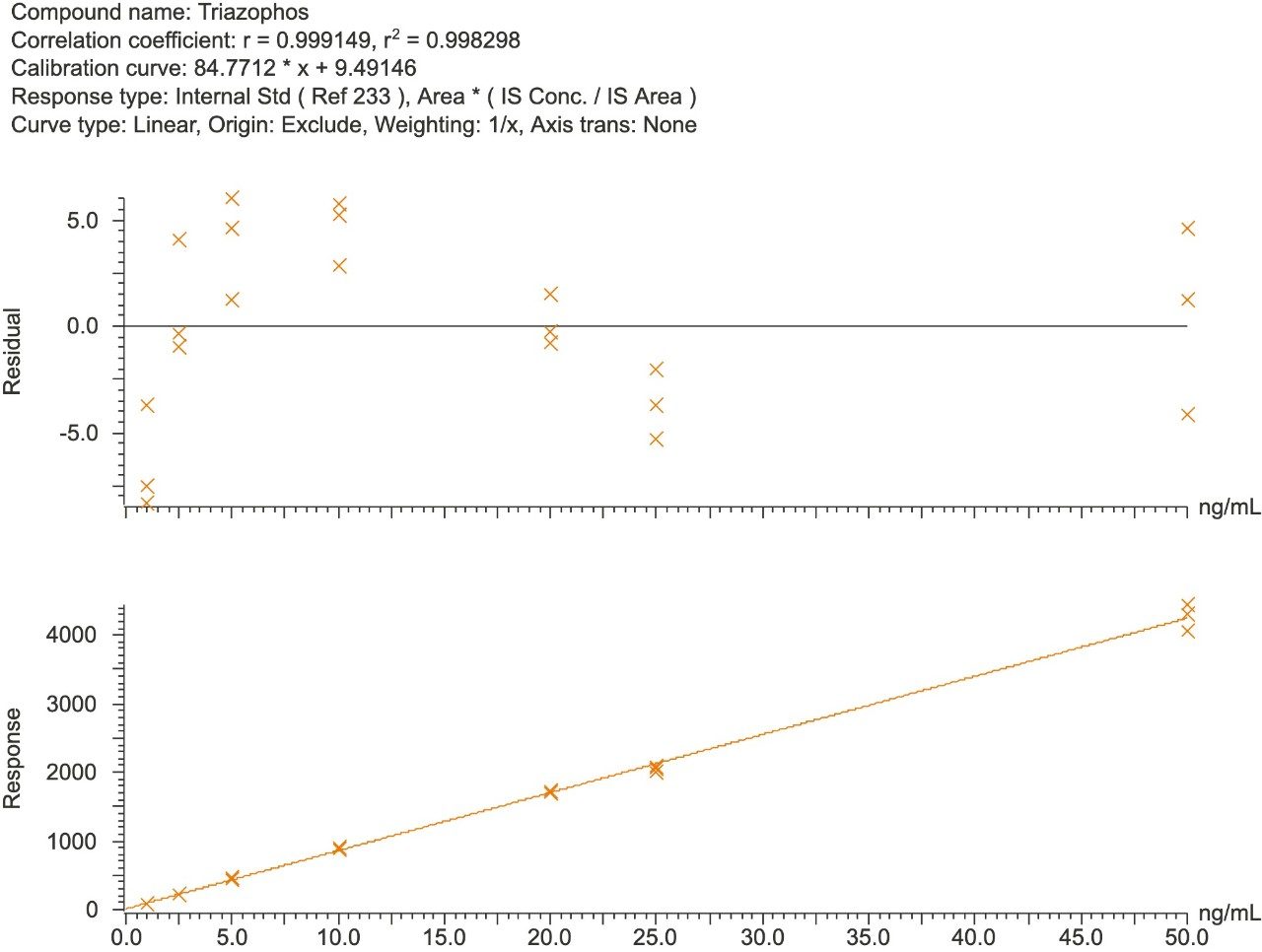
Solvent and matrix match spiked calibration (MMS) curves were prepared at concentrations that equated to the range 1 ppb to 50 ppb (i.e. 0.001 to 0.05 mg/kg of okra) and injected in triplicate. The majority of the compounds showed linearity with R2 values greater than 0.99 in both the solvent and MMS curves. Ethoxyquin, milbemectin A3, and A4, oxadiazon, spiromesifen, and terbufos showed R2 values greater than 0.970 for both solvent and MMS curves. However, fipronil, phorate, and thiabendazole showed R2 values greater than 0.970 in MMS curves only. Figures 2 and 3 show calibration curves and residuals for an example compound (triazophos) in solvent and matrix respectively.
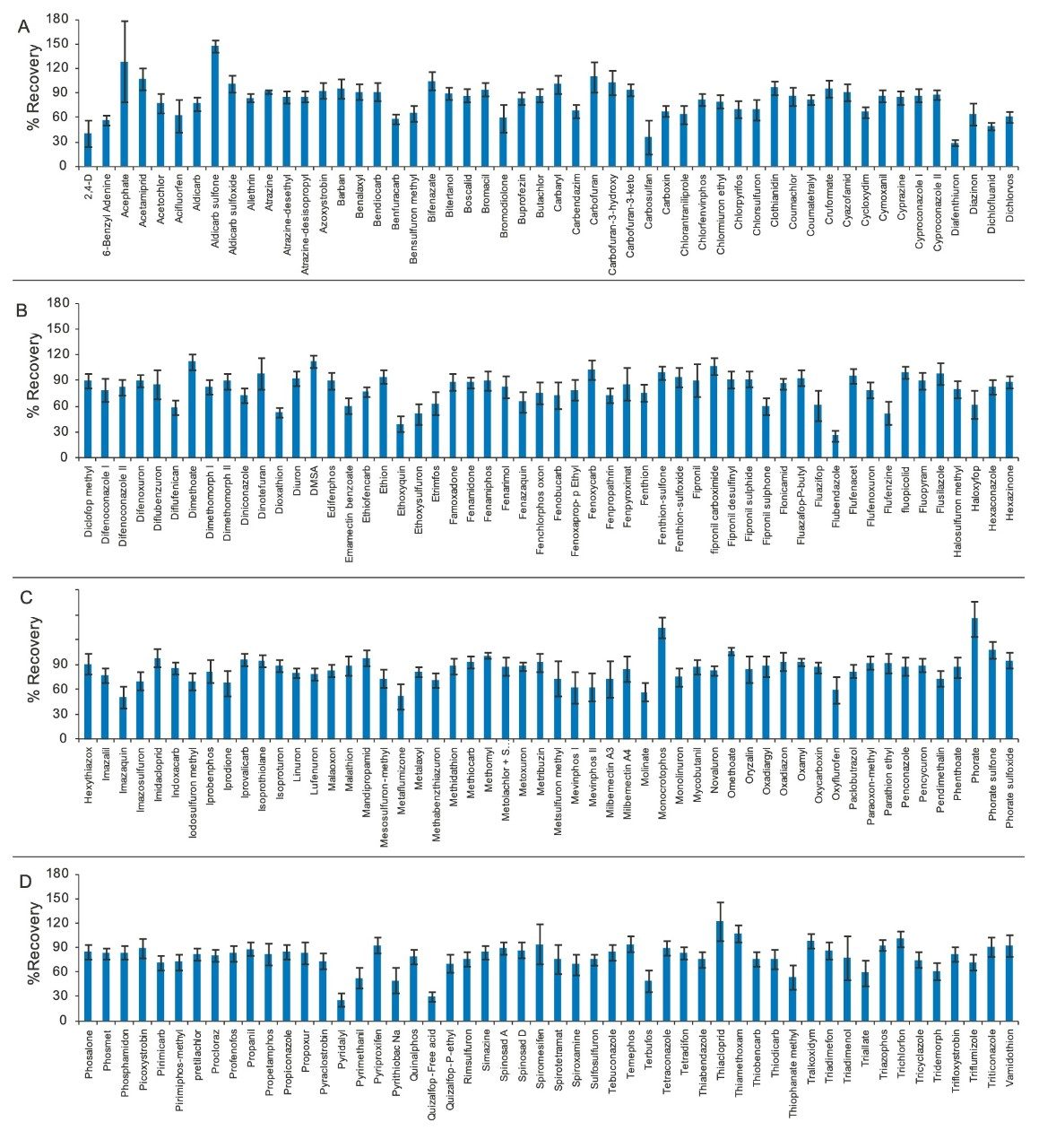
To evaluate the recovery, accuracy, and precision of the method, studies were carried out on spiked samples. Okra samples were pre-spiked with all the pesticides at 10 ppb (0.01 mg/kg) in triplicate, extracted, and quantified against the MMS calibration curve. Recoveries were calculated using TargetLynx Software. The recoveries reported are without any internal standard correction. As shown in Figure 4 (A, B, C, and D), recoveries for all of the pesticides ranged from 25% to 150%. Relative standard deviations (RSDs, shown as error bars in Figure 4) for most compounds were <20%. The RSDs for 34 compounds were found to be higher than 20%. Use of an internal standard would be likely to significantly improve repeatability for those analytes.
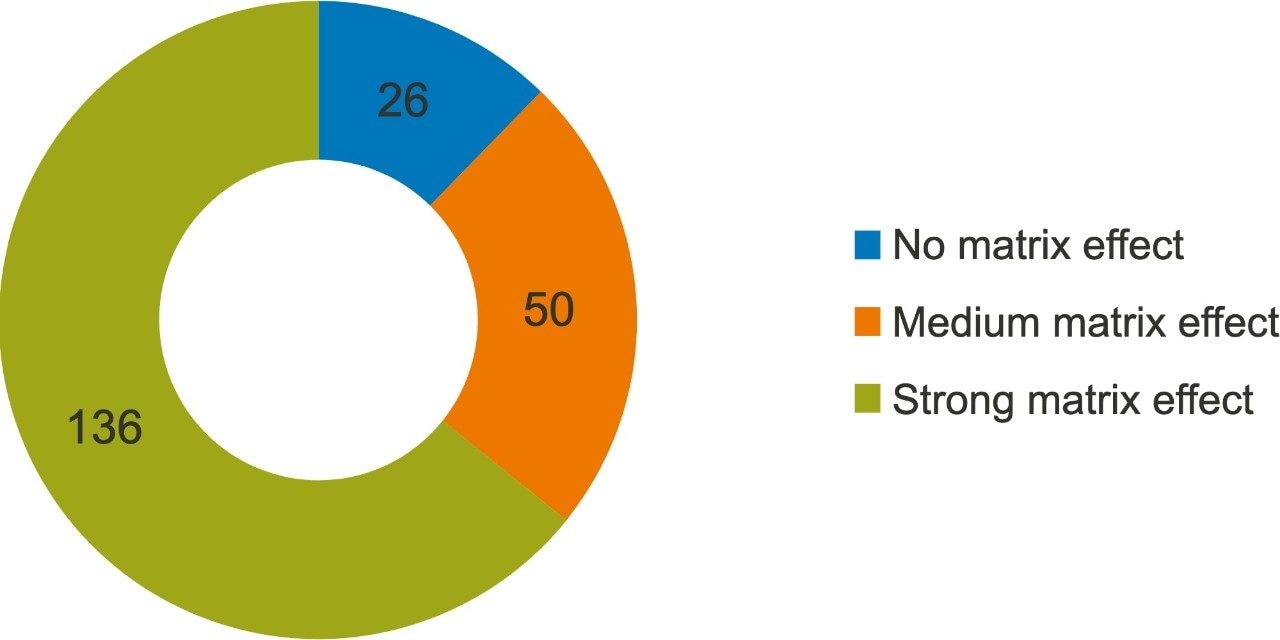
Matrix effects for all of the pesticides were calculated by taking the ratio of the slope of the MMS calibration curve to the slope of solvent calibration curve. A percent variation of + 20% was considered as no matrix effect as this variation is close to the repeatability values.6 Values between + 20% to + 50% were considered as a medium matrix effect, and a strong matrix effect was considered to be values greater than + 50%.7 Figure 5 shows levels of the matrix effect that were observed in the analysis of okra for all pesticides. A strong matrix effect was observed for the majority of compounds, demonstrating that the analysis of okra samples poses a challenge in regards to high matrix complexity. Even with these high matrix effects, all compounds can easily be detected at legislative limits and quantified using the matrix-matched calibration curve.
To further understand the impact of co-eluting matrix components that can compete with an analyte of interest during the ionization process, RADAR technology enables the simultaneous acquisition of full spectrum data during quantitative MS/MS analysis. Figure 6 shows an example of the use of RADAR technology. In Figure 6A, the base peak intensity (BPI) chromatogram from the full-scan background data for the okra sample is shown. At 5.08 minutes, close to the retention time of dimethoate (Figure 6B and 6C), high matrix interference was observed. The spectrum at 5.08 minute showed an intense ion at m/z 217.1 (Figure 6D).
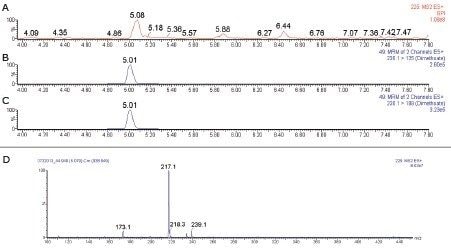
This interferent potentially has a large impact on the detection of dimethoate and a 48% ion suppression effect was observed for dimethoate. In the case of aldicarb, however, matrix interference was minimal (0.4%) and the RADAR data (Figure 7) showed no evidence of interferences at the retention time of aldicarb (6.13 minutes). The spectrum at the retention time of aldicarb has been expanded and zoomed in the inset (Figure 7D), clearly demonstrating that there was a much higher response from co-extracted matrix ions at the retention time of dimethoate compared to aldicarb. These data clearly demonstrate the usefulness of RADAR technology in assessing the matrix background and its potential effect on ion enhancement or suppression.

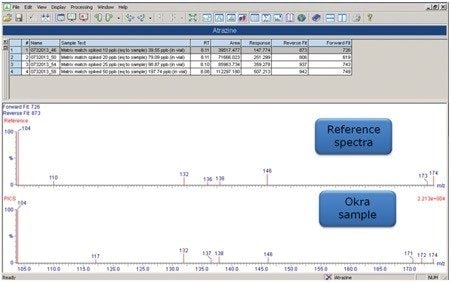
In complex matrices, situations arise where closely-related compounds such as metabolites or matrix interferences show responses for the target compounds of interest, even in MRM mode. This can lead to ambiguity and may require an additional qualitative experiment. An alternative is to employ a product ion confirmation scan (PICs) within the quantitative MRM experiment. PICs can be used to confirm peak identity through automatic acquisition of an MS/MS spectrum after the apex of the peak has eluted. PICs, in combination with TargetLynx, provides additional confirmation of the compounds of interest through comparison of the acquired MS/MS spectrum to a reference spectrum. Figure 8 shows the TargetLynx results from the comparison of the atrazine MS/MS spectrum obtained from PICS in an okra sample versus the reference spectrum, which was obtained from MS/MS analysis of the standard in solvent.
720004789, September 2013