This application note presents the use of an exact mass UPLC-MS/MS analytical method for structural characterization of 21-nt-long RNA.
High mass accuracy and resolution
RNA interference (RNAi) mechanism plays a fundamental role in post-transcriptional gene silencing. With a knowledge of sequence, gene silencing experiments are now routinely performed using ~21 nucleotide (nt) long synthetic RNAi probes, which are also being developed as therapeutics.
Synthetic RNAi oligonucleotides (siRNA) need to be purified to avoid an off-target silencing of undesirable genes. RNAi drugs need to be well-characterized to satisfy regulatory requirements and minimize possible adverse implications for safety and efficacy.
The high-resolution chromatographic capabilities of Waters UltraPerformance LC (UPLC) Technology coupled with mass spectrometry analysis provides a powerful tool for the analysis of biopharmaceutical drugs such as siRNA oligonucleotides. An important part of oligonucleotide characterization is sequencing, which can be performed using a selective enzyme or chemical. Methods employing MS/MS fragmentation are more generic and faster, and can be used for modified oligonucleotides, which are often resistant to enzymatic cleavage. In order to obtain structural information for the whole 21 nt RNAi, the molecules should be efficiently ionized and produce sequence-related ions during MS/MS using proper mass accuracy and resolution.
This application note presents the use of an exact mass UPLC-MS/MS analytical method for structural characterization of 21-nt-long RNA. The method is highly useful for confirmatory sequencing of siRNA base therapeutic compounds.
Complementary RNA strands of 21 nt length, upper strand 5' -UCG UCA AGC GAU UAC AAG GTT -3', and lower strand 5' -CCU UGU AAU CGC UUG ACG ATT -3', were reconstituted separately in 0.1 M triethylamine acetate (TEAA). A distribution of synthetic byproducts was detected along with the upper and lower strands by UPLC-MS. The sample concentration used for the UPLC-MS analysis was 30 pmol/μL.
UPLC-MS analysis was performed as described previously.1 The Waters ACQUITY UPLC System was used with an ACQUITY UPLC Oligonucleotide Separation Technology (OST) C18 Column, 1.7 μm, 2.1 x 50 mm (PN 186003949), for analysis. Mass spectrometry parameters were chosen in order to achieve maximum declustering of TEA adducts without compromising the intensity of the precursor ion. The Waters SYNAPT HDMS Mass Spectrometer was operated in time-of-flight (TOF) MS mode.
Collision-induced dissociation for selected ions was performed with a collision energy ramp from 25 to 55 V. The extent of MS/MS fragmentation was manipulated by selecting appropriate charge states (-3 to -6) and varying the collision energy ramp. Product ion spectra were acquired over the range of 500 to 7000 m/z at a rate of 1 scan/sec. External calibration in negative ion mode was performed with cesium iodide. MaxEnt 3 Software was used for spectral deconvolution prior to data analysis.
|
LC system: |
Waters ACQUITY UPLC System |
|
Column: |
ACQUITY UPLC OST C18 1.7 μm, 2.1 x 50 mm |
|
Column temp: |
60 °C |
|
Injection volume: |
5 μL |
|
Flow Rate: |
0.2 mL/min |
|
Mobile phase A: |
15 mM TEA, 400 mM HFIP |
|
Mobile phase B: |
50% A, 50% methanol |
|
Gradient: |
20 to 40% B in 10 min |
|
MS system: |
Waters SYNAPT HDMS System |
|
Capillary: |
2.7 V |
|
Sampling cone: |
31 V |
|
Extraction cone: |
3 V |
|
Source temp.: |
120 °C |
|
Desolvation temp.: |
300 °C |
|
Desolvation gas flow: |
500 L/h |
|
Trap collision energy: |
6 V |
|
Transfer collision energy: |
4 V |
|
Mass resolution: |
~9,000 in V mode (FWHH) |
|
LockMass: |
CsI 10 mg/mL (water-isopropanol, 1:1), 5 μL/min flow rate, 1 sec scan time, 30 sec frequency, set mass 1685.765 m/z (Cs6I7-) |
The collision energy profile affects the extent of MS/MS fragmentation; energy values need to be adjusted for the specific analyte of interest. Typically, the most abundant charge state is selected for MS/MS fragmentation, which for the 21-nt-long species is between -5 and -3 in the 1500 to 2000 m/z range. A collision energy ramp from 25 to 45 V was suitable to generate structurally-useful fragments for 21 nt RNA.
In general, in order to obtain reasonable signal-to-noise (S/N) for MS/MS fragmentation, a minimum signal of 500 ion counts is desirable for the main component or a contaminant in a total ion chromatogram (TIC).
Complementary RNA 21 nt strands were separately injected onto the UPLC-MS/MS system. The chromatogram revealed shorter oligonucleotide peaks (products of 5' hydrolysis) that are wellresolved from the original target 21 nt (Figure 1). Components were assigned based on their exact mass measurement, providing partial sequence verification.1
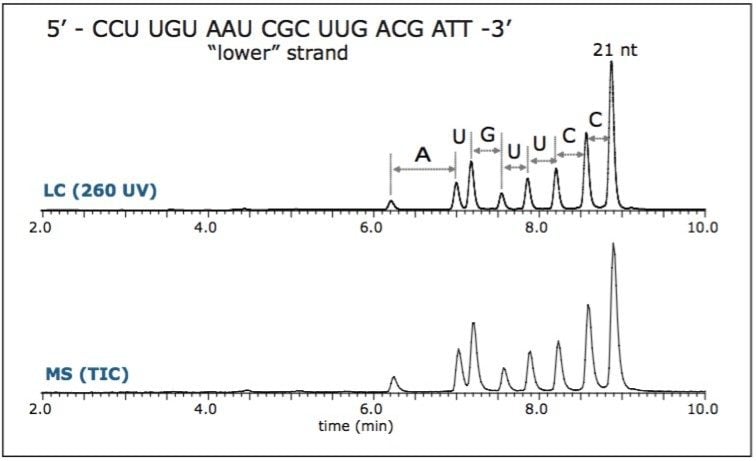
In order to verify the structure of the whole oligonucleotide, the precursor ion, [M-4H]4-, from the 21 nt lower strand was isolated and fragmented (Figure 2). The predominant characteristic fragments were complementary c and y-ions with low intensity [a-B] and w ions (nomenclature is shown in Figure 3). These sequence ions resulting from 5'-P-O cleavage are prevalent among the MS/MS fragments of RNA oligonucleotides, in contrast to those produced from DNA, which are almost exclusively [a-B] and w ions from 3'-C-O cleavage. This is due to the absence of the 2'-hydroxyl group in DNA molecule.
Internal fragments, produced by double cleavage of the backbone as well as a neutral loss of cytosine from c-ion (Table 1) were significantly less abundant and did not contribute to structurally-useful peak assignments. MS/MS analysis of the upper 21 nt RNA strand through ramping of collision energy from 30 to 50 V yielded similar results (Figure 2).
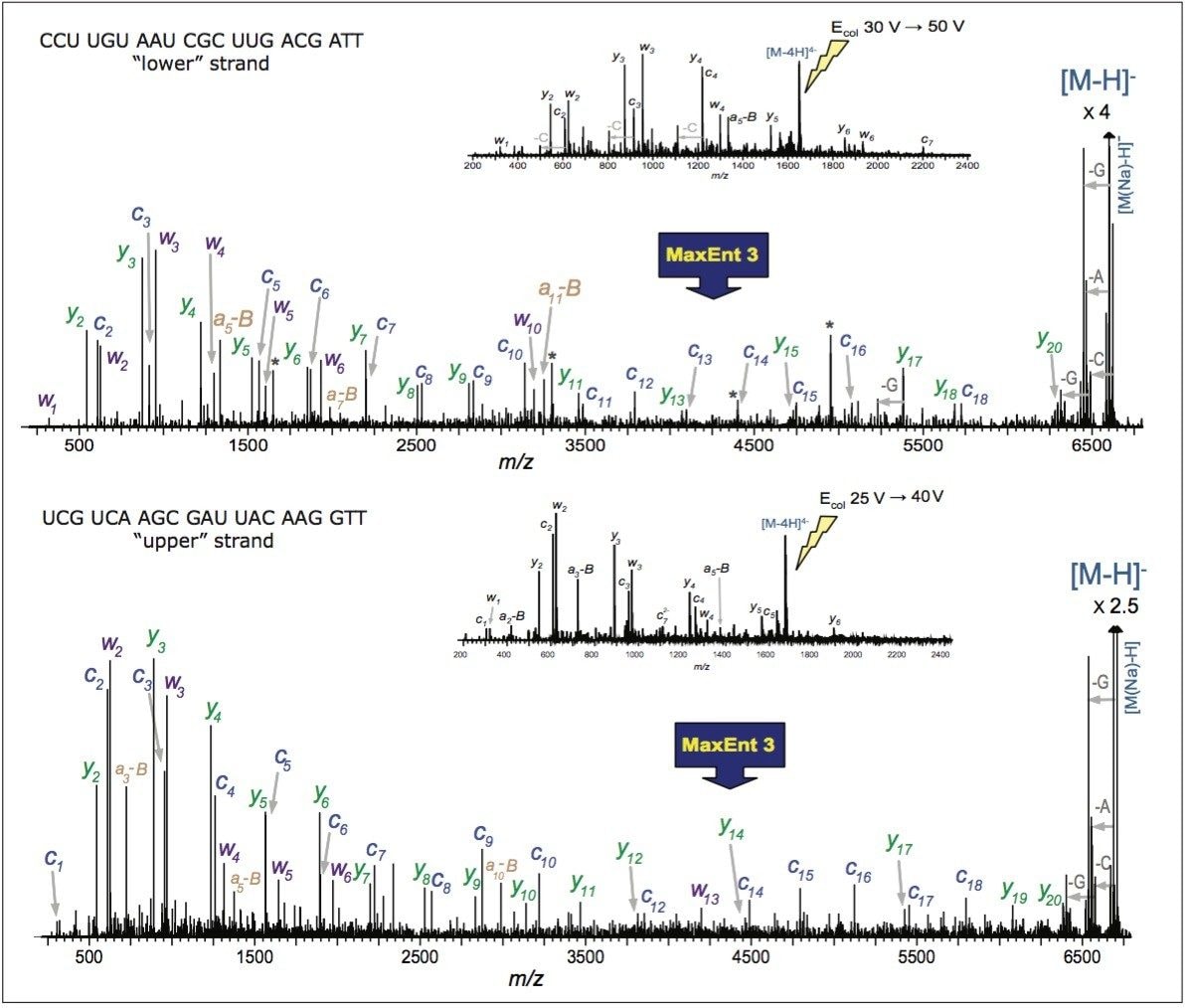
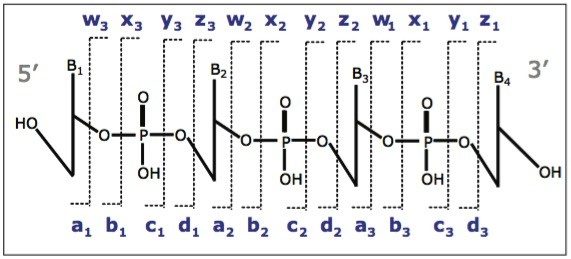
Of all the fragment ions in the MS/MS spectra of [M-4H]4-, 21 nt RNA were represented in several charge states (Table 1). Deconvolution of the MS/MS spectrum to singly-charged ions was performed using MaxEnt 3 software and significantly reduced the spectral complexity, simplifying the MS/MS data interpretation.
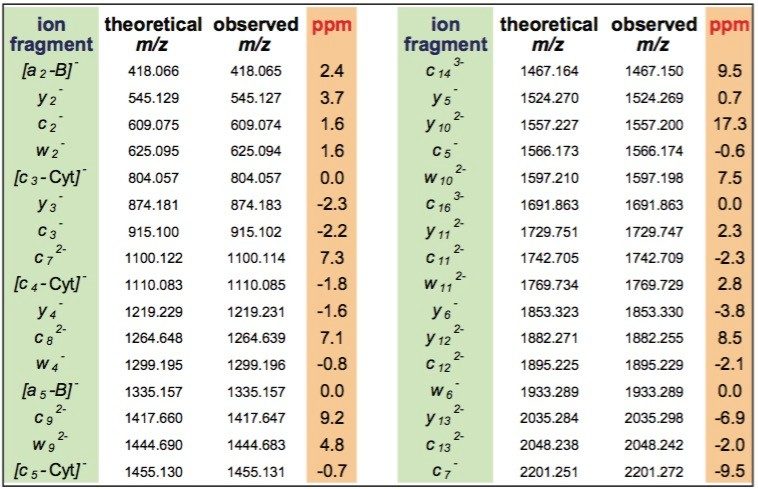
The deconvoluted spectra was sufficient to interpret the entire siRNA sequence (Figure 2). A few peaks corresponding to A, G, and C gas-phase nucleobase losses from the 21 nt molecular ion were also detected. The LockMass calibration, yielding a mass accuracy below 10 ppm, is highly useful for manual data interpretation (Table 1). Mass resolution of triple quadrupole mass spectrometers and ion trap instruments may not be sufficient for unambiguous assignment of characteristic fragments and distinguishing among multiply-charged peaks and other products of oligonucleotides' complex fragmentation.
The automation of MS/MS enables the analysis time to be reduced by hours, if not days, and to decrease the cost of sample analysis. In addition, UPLC enables fast separation of RNA species of interest; multiple oligonucleotide strands can be analyzed by MS and MS/MS in a single analysis. The ability of UPLC to resolve target oligonucleotides from their truncated products is highly desirable for siRNA metabolism studies.
A reliable sequencing method via exact mass UPLC-MS/MS was developed for the structural characterization of siRNA oligonucleotides.
This efficient confirmatory sequencing method is very useful for sequence verification of therapeutic oligonucleotides required by the U.S. FDA for biotherapeutic compounds. The method is potentially applicable also for de novo sequencing of unknown impurities. It is expected that this MS/MS approach is suitable for chemically-modified oligonucleotides that are difficult to digest with exonucleases (ladder sequencing).
720002869, December 2008