For research use only. Not for use in diagnostic procedures.
This application note highlights the performance characteristics of the ACQUITY UPLC M-Class System operating at nanoscale flow rates, illustrating its utility in analyzing complex, tryptic-peptide mixtures.
Operating at nanoscale flow rates in proteomics applications, the ACQUITY UPLC M-Class System confers these benefits:
Nanoscale chromatography is established as the method of choice in bottom-up, or shotgun, proteomics experiments. It achieves superior sensitivity, compared with higher flow-rate separations, because of the reduced dilution effects of peptides of low stoichiometry present within the sample. When this scale of chromatography is coupled to a QToF mass spectrometer operating in the data-independent (DIA), data-dependent (DDA), or multiple-reaction-monitoring (MRM) acquisition mode, its higher peak-capacity separations yield enhanced results, namely, higher levels of protein/peptide identifications and lower detection and quantitation limits.
The ACQUITY UPLC M-Class System offers direct-flow separation for flow rates ranging from nanoscale to microscale, with an upper limit of 15,000 psi operating pressure. This higher pressure limit, compared with the nanoACQUITY UPLC System, permits the use of longer columns packed with sub-2-μm particles, for maximum separation efficiency. It also enables higher flow rates when shorter columns are used for high-throughput analyses.
This application note demonstrates the salient performance characteristics of the ACQUITY UPLC M-Class System, reporting typical results from DIA HDMSE acquisitions of a complex, tryptic-peptide sample.
|
LC conditions |
|
|---|---|
|
LC system: |
ACQUITY UPLC M Class |
|
Trapping column: |
ACQUITY UPLC M-Class Symmetry C18, 100Å, 5 μm, 180 μm x 20 mm, 2G, V/M Trap Column (p/n 186007496) |
|
Trapping conditions: |
Isocratic delivery of aqueous 0.1% formic acid, at 5 μL/min for five minutes |
|
Analytical column: |
ACQUITY UPLC M-Class HSS T3, 100Å, 1.8 μm, 75 μm x 250 mm Column (p/n: 186007474) |
|
Column temperature: |
35 °C |
|
Elution flow rate: |
300 nL/min |
|
Mobile phase A: |
Aqueous 0.1% formic acid |
|
Mobile phase B: |
0.1% formic acid in acetonitrile |
|
Gradient: |
1% to 30% B linear gradient, in 30 or 90 min |
|
Instrument: |
SYNAPT G2-Si |
|
Acquisition mode: |
MSE and HDMSE modes |
|
Mass range: |
50 to 2000 Da |
|
Integration time: |
0.2s (MSE) or 0.5s (HDMSE) |
ProteinLynx Global SERVER (PLGS) Software
Spotfire (Tibco, Boston, Massachusetts, USA)
As measured by both retention-time reproducibility and peak width, chromatographic performance is a central factor in the analysis of complex mixtures. Data acquisitions that rely on quantitative, label-free strategies also rely on retention-time reproducibility to identify and quantify analytes from multiple injections. Furthermore, narrow peak widths, which result from operating at UPLC pressures and from using columns packed with smaller particles, contribute to higher peak-capacity separations. More effective separations augment the data-processing algorithms used for label-free quantitation. Such separations better resolve ambiguity, and they elucidate closely associated precursors and fragment ions because of less interference from co-elutions.
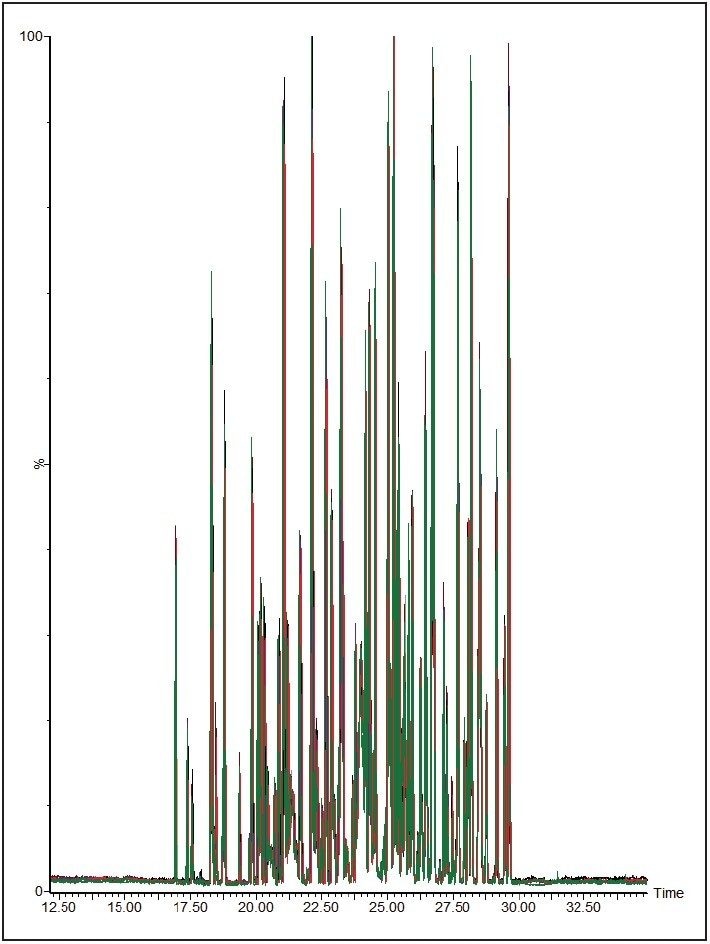
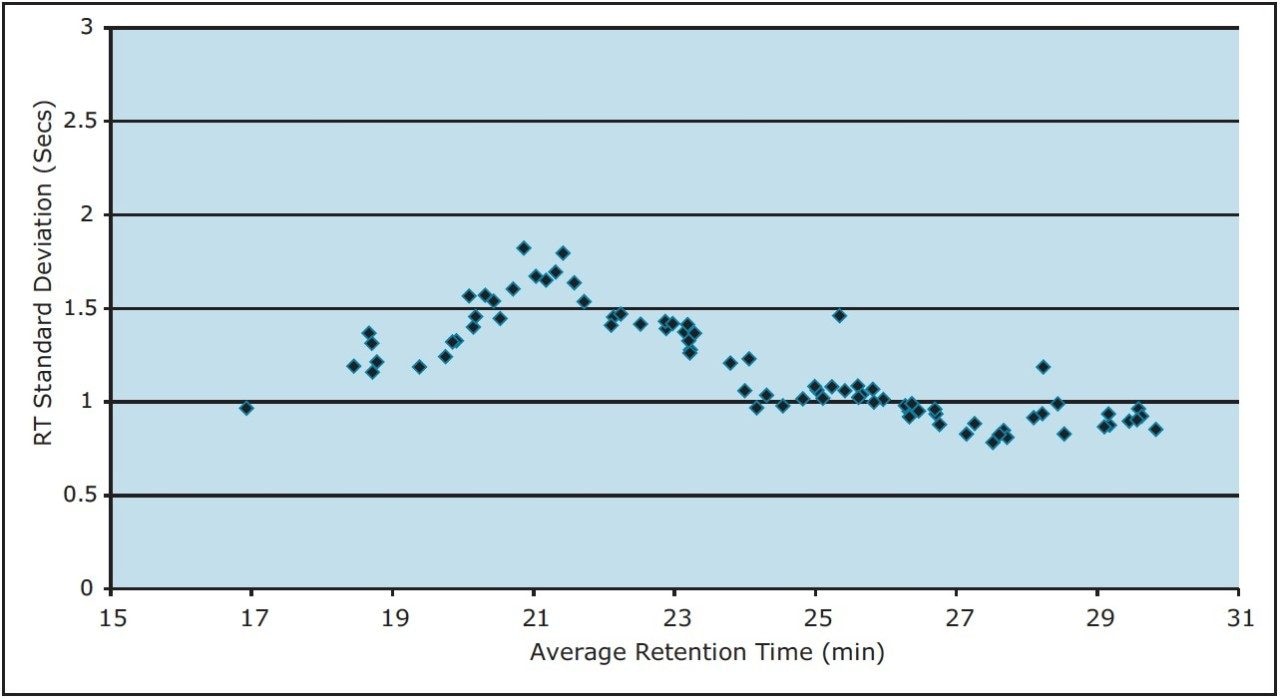
To measure the retention-time reproducibility and peak widths of the ACQUITY UPLC M-Class System, six repeat injections of MPDS Mixture 1 were made and the digest separated using a 30-minute gradient. Their chromatograms appear in Figure 1. Figure 2 shows standard deviations of the 84 most intense peptides, compared with their average retention time. All of these peptides show an excellent standard deviation of less than or equal to 2 seconds. Figure 3 shows the frequency distribution of the chromatographic peak widths of the same peptides. Most peaks elute with a full-width at half-maximum (FWHM) width of 3.0 to 3.6 seconds or a solvent volume of 18 nL, equating to a theoretical peak capacity of 213 for this separation.
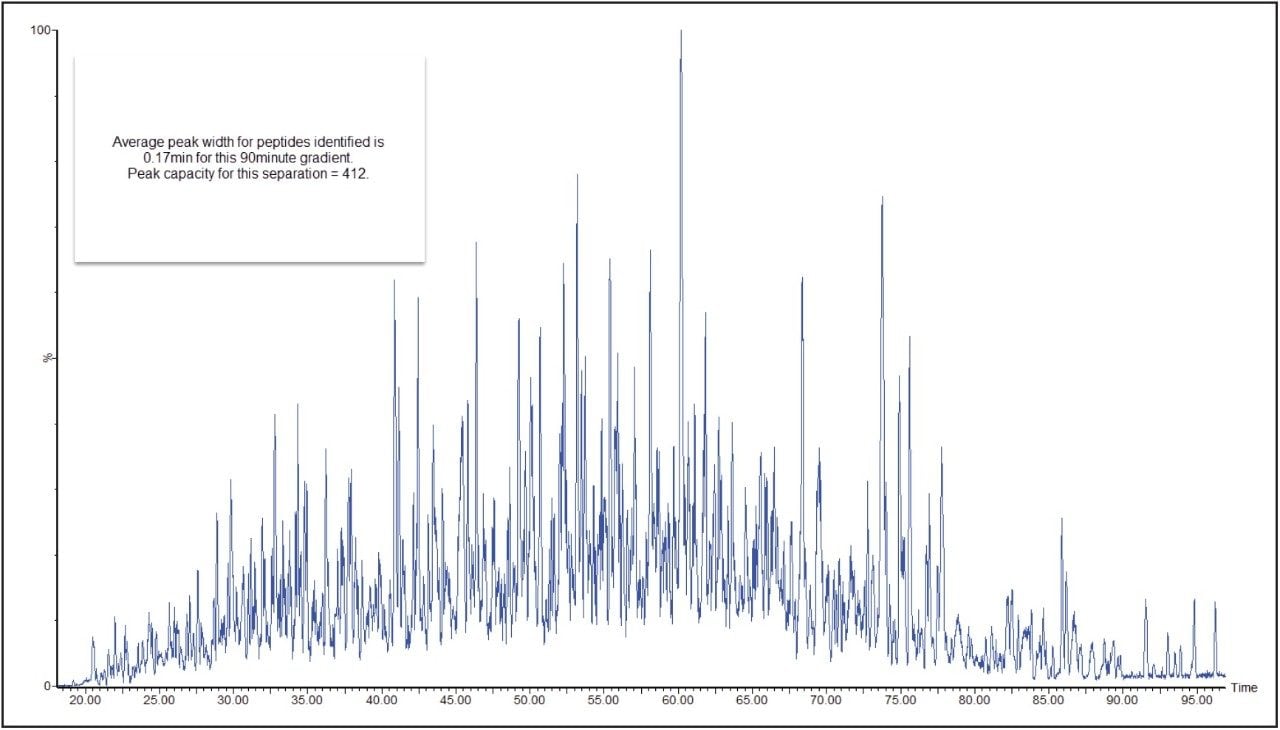
In a DIA HDMSE acquisition, fragments correlate with precursors according to their ion mobility as well as elution profiles. Samples analyzed in this way lead to higher protein identifications, compared with Tof-only MSE – that is, without ion-mobility separation. Figure 3 shows a typical chromatogram from the analysis of 100 ng of a relatively complex tryptic digest of cytosolic E. coli. Peaks elute in this separation with an average FWHM width of 0.17 min, leading to a theoretical peak capacity of 412. When processed in PLGS software, protein identifications exceeded 1100, with more than 20,000 peptides identified, as shown in Figure 4.
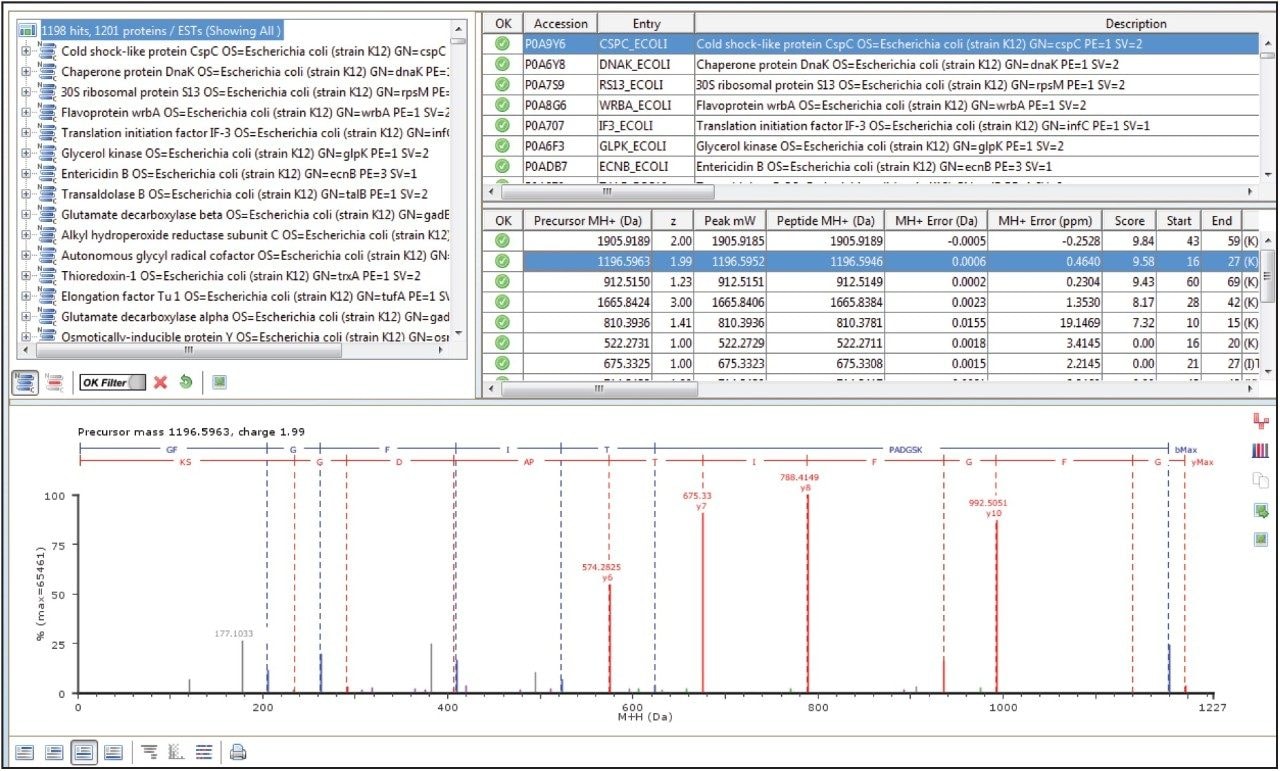
This application note highlights the performance characteristics of the ACQUITY UPLC M-Class System operating at nanoscale flow rates, illustrating its utility in analyzing complex, tryptic-peptide mixtures. Retentiontime reproducibility, with standard deviations of less than 2 seconds, is achieved with peak widths for most peptides of 3.6 seconds FWHM over a short gradient. For analyses of complex mixtures, the system coupled to a SYNAPT G2-Si Mass Spectrometer operating in HDMSE mode has been shown to achieve excellent protein identification rates.
720005244, March 2015