Oligo Mapping of sgRNA Digests: Leveraging Xevo MRT Mass Spectrometer Performance and Streamlining Data Analysis
Catalin Doneanu, Alexandre F Gomes, Tatiana Johnston, Chris Preston, Matt Gorton, Bala Addepalli, Scott Berger, Ying Qing Yu
Waters Corporation, Milford, MA, United States
Published on September 15, 2025
Abstract
Recent advances in liquid chromatography-mass spectrometry (LC-MS) and bioinformatics technologies have enhanced our detailed understanding of therapeutic RNA molecules. This application note presents a compliance-ready workflow for digested single-guide RNA (sgRNA) oligo mapping. The workflow incorporates sample preparation using the RapiZyme™ MC1 ribonuclease, UPLC™-MSE data acquisition on the Xevo™ MRT Mass Spectrometer and automated data analysis on the waters_connect™ MAP Sequence App (ver 1.0). The Xevo MRT Mass Spectrometer combines cutting-edge multi-reflecting time-of-flight technology with hybrid quadrupole time-of-flight (QTOF) capabilities, delivering sub-ppm mass accuracy that ensures confident identification of digested RNA products. In this study, complete (100%) unique (non-ambiguous) sequence coverage of an sgRNA was achieved based on the unique accurate masses of the sgRNA digestion products. The CONFIRM Sequence App was also employed to assign isomeric digestion products by validating their sequence from elevated energy fragmentation data gathered in the same run used for digestion product mapping.
Benefits
- Confident characterization of sgRNA digestion products with sub-ppm mass accuracy of oligonucleotide precursors, outstanding MS resolution (~100,000), and sensitivity at high acquisition speed
- A compliance-ready informatics workflow featuring the waters_connect MAP Sequence App (ver 1.0) streamlines oligonucleotide mapping of sgRNA digests
- RapiZyme MC1, a new ribonuclease - offers unique cleavage specificity and the opportunity to generate overlapping digestion products, enabling complete (100%) sequence coverage for sgRNAs
Introduction
Single guide RNAs (sgRNAs) were first described in 20121 when two RNA molecules essential for CRISPR-Cas9-mediated DNA cleavage – the trans-activating CRISPR RNA (tracrRNA), which serves as a scaffold for the Cas9 nuclease, and the CRISPR RNA (crRNA), responsible for DNA target recognition - were fused into a single RNA construct. This fusion created the sgRNA, a critical component of the CRISPR-Cas9 gene editing system.1–2 The sgRNA molecule directs the Cas9 nuclease to introduce precise double-stranded breaks in DNA, enabling targeted genetic modifications.
Since its discovery, which was honored with the Nobel Prize in Chemistry, CRISPR technology has transformed gene editing applications. Beyond basic research, it has been adapted for rapid diagnostic tools, such as COVID-19 tests developed during the 2020 global pandemic3, and holds promise for therapeutic interventions in genetic diseases, cancer, and infectious diseases. As sgRNAs are typically synthesized via solid-phase oligonucleotide synthesis, their analytical characterization requires confirmation of both the intact molecular weight4 and the sequence verification to ensure functional accuracy.5
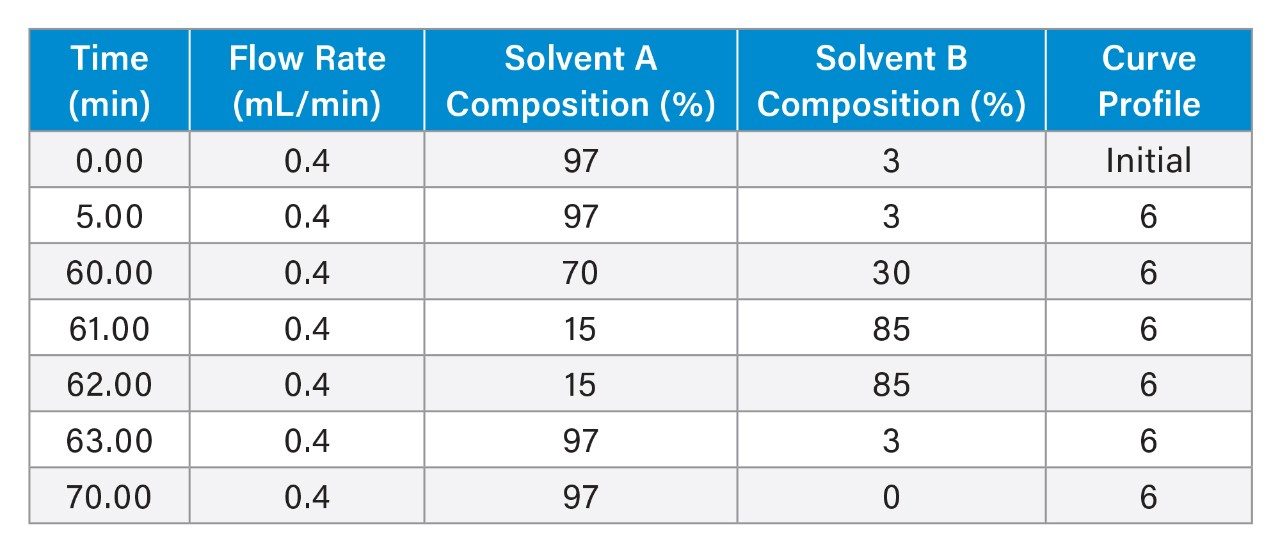
Traditional LC-MS workflows for sgRNA digest oligo mapping have been labor-intensive and time-consuming, relying heavily on manual data analysis and interpretation. Recently6, a UPLC-MS and informatics workflow designed for automated sgRNA sequence mapping was presented following enzymatic digestion with a panel of ribonucleases, including RNase T1, hRNAse 4 and two RNase T2 enzymes: RapiZyme MC1 and Cusativin. This application note extends the effectiveness of this workflow (Figure 1) when implemented on a Xevo MRT Mass Spectrometer (Figure 1), while focusing specifically on MC1 digestion of the Waters™ sgRNA LC-MS standard, where complete unique digested product coverage was obtained.
Experimental
Reagents and Sample Preparation
N,N-diisopropylethylamine (DIPEA, 99.5% purity, catalog number 387649-100ML) was purchased from Millipore Sigma (St Louis, MO) and 1,1,1,3,3,3-hexafluoro-2-propanol (IonHance HFIP, p/n:186010781) was obtained from Waters (Milford, MA). Methanol (LC-MS grade, catalog number 34966-1L) and acetonitrile (LC-MS grade, catalog number 34967-6XL) were obtained from Honeywell (Charlotte, NC). HPLC grade Type I deionized (DI) water was purified using a Milli-Q system (Millipore, Bedford, MA). Mobile phases were prepared fresh daily. Ultrapure nuclease-free water (catalog number J71786.AE) for sgRNA digestions was purchased from Thermo Fisher Scientific (Waltham, MA).
The Waters sgRNA LC-MS standard (p/n:186011357) is a 100-mer single-guide RNA (sgRNA), encoding specifically for the mouse GATA2 transcription factor. Its sequence is: 5’ - C*U*U* CAA CCA UCU CGA CUC GCG UUU UAG AGC UAG AAA UAG CAA GUU AAA AUA AGG CUA GUC CGU UAU CAA CUU GAA AAA GUG GCA CCG AGU CGG UGC U*U*U* U-3’. It contains a 2’-OMe modification on its first three 5’ nucleotides (C*U*U*, denoted as the WYY sequence shown in Figure 5), as well as on its last three 3’ nucleotides (U*U*U*, sequence YYY in Figure 5). The asterisk indicates that all these six nucleotides are phosphorothioated. A 50 µM stock solution of the Waters sgRNA LC-MS standard was prepared using nuclease-free water.
RapiZyme MC1 (Waters p/n:186011190, 10000 units/tube) is a novel RNA digestion enzyme recently introduced by Waters Corporation.6-7 10.6 µL of 50 µM Waters sgRNA standard (p/n:186011357) was denatured at 90 oC for 2 minutes in a buffer containing 200 mM ammonium acetate (catalog no AM9071-500ML, Thermo Fisher, Waltham, MA) pH 8.0. The sample was cooled on ice and microcentrifuged to collect the sample droplets. After adding 100 units of digestion enzyme (2 µL of RapiZyme MC1) and 7.4 µL of nuclease-free water to obtain a final volume of ~ 20 µL, the sgRNA was digested at 30 ºC for 30 minutes in an Eppendorf thermomixer. The digest was then analyzed immediately by LC-MS using 5 µL injections.
All datasets were acquired with waters_connect (ver 4.1.0.17) and subsequently processed using the MAP Sequence App (ver 1.0), assisted by the mRNA Cleaver and Coverage Viewer MicroApps.
LC Conditions
|
LC-MS system: |
Xevo MRT (multi-reflecting time-of-flight) Mass Spectrometer coupled with ACQUITY™ Premier UPLC (Binary) System |
|
Column: |
ACQUITY Premier Oligonucleotide BEH™ C18 Column 300 Å, 1.7 µm, 2.1 x 150 mm, (p/n:186010541) |
|
Column temperature: |
70 °C |
|
Flow rate: |
400 µL/min |
|
Mobile phases: |
Solvent A: 0.1% DIPEA (N,N-diisopropylethylamine), 1% HFIP (1,1,1,3,3,3-hexafluoroisopropanol) in DI water, pH 8.5 Solvent B: 0.0375% DIPEA, 0.075% HFIP in 65% ACN |
|
Sample temperature: |
8 °C |
|
Sample vials: |
QuanRecovery™ MaxPeak™ HPS vials (p/n:186009186) |
|
Injection volume: |
5 µL |
|
Wash solvents: |
Purge solvent: 10% methanol in DI water Sample manager wash solvent: 50% MeOH Seal wash: 10% methanol in DI water |
Gradient Table
MS Conditions
|
MS system: |
Xevo MRT (multi reflecting time-of-flight) QTOF Mass Spectrometer |
|
Ionization mode: |
ESI(-) |
|
Acquisition mode: |
MSE |
|
Acquisition rate: |
2 Hz |
|
Capillary voltage: |
1.5 kV |
|
Cone voltage: |
40 V |
|
Source offset: |
10 V |
|
Source temperature: |
120 oC |
|
Desolvation temperature: |
550 °C |
|
Cone gas flow: |
0 L/h |
|
Desolvation gas flow: |
1000 L/hr |
|
TOF mass range: |
50–4000 (MSE acquisition) |
|
Low energy CE: |
6 V |
|
High energy CE ramp: |
15 to 25 V |
|
Lock-mass: |
50 pg/µL Leu Enk in 0.1% formic acid, 50% ACN |
|
Data acquisition: |
waters_connect version 4.1.0.17 mRNA Cleaver Micro App version 1.1.0 MAP Sequence App version 1.0 Coverage Viewer Micro App version 2.0.0 |
Results and Discussion
An informatics workflow (Figure 1) featuring the waters_connect MAP Sequence App (version 1.0) was used to facilitate automated data processing of UPLC-MSE datasets acquired following the enzymatic digestion of an sgRNA standard with RapiZyme MC1.6–7 The informatics processing workflow consisted of three steps:
1. In-silico digestion: The mRNA Cleaver MicroApp was used to generate predicted, in-silico digested oligonucleotide digestion products, based on the target RNA sequence.
2. Data processing and mass matching: The waters_connect MAP Sequence App processed the UPLC-MSE data, matching the predicted neutral monoisotopic masses of digested oligonucleotides to the experimentally acquired MS1 data.
3. Sequence coverage visualization: The resulting sequence coverage for the RNA digest was summarized and visualized using the Coverage Viewer MicroApp.
All data acquisition and processing were conducted within the waters_connect Informatics Platform to ensure regulatory compliance.
Digested Oligo Mapping of sgRNA
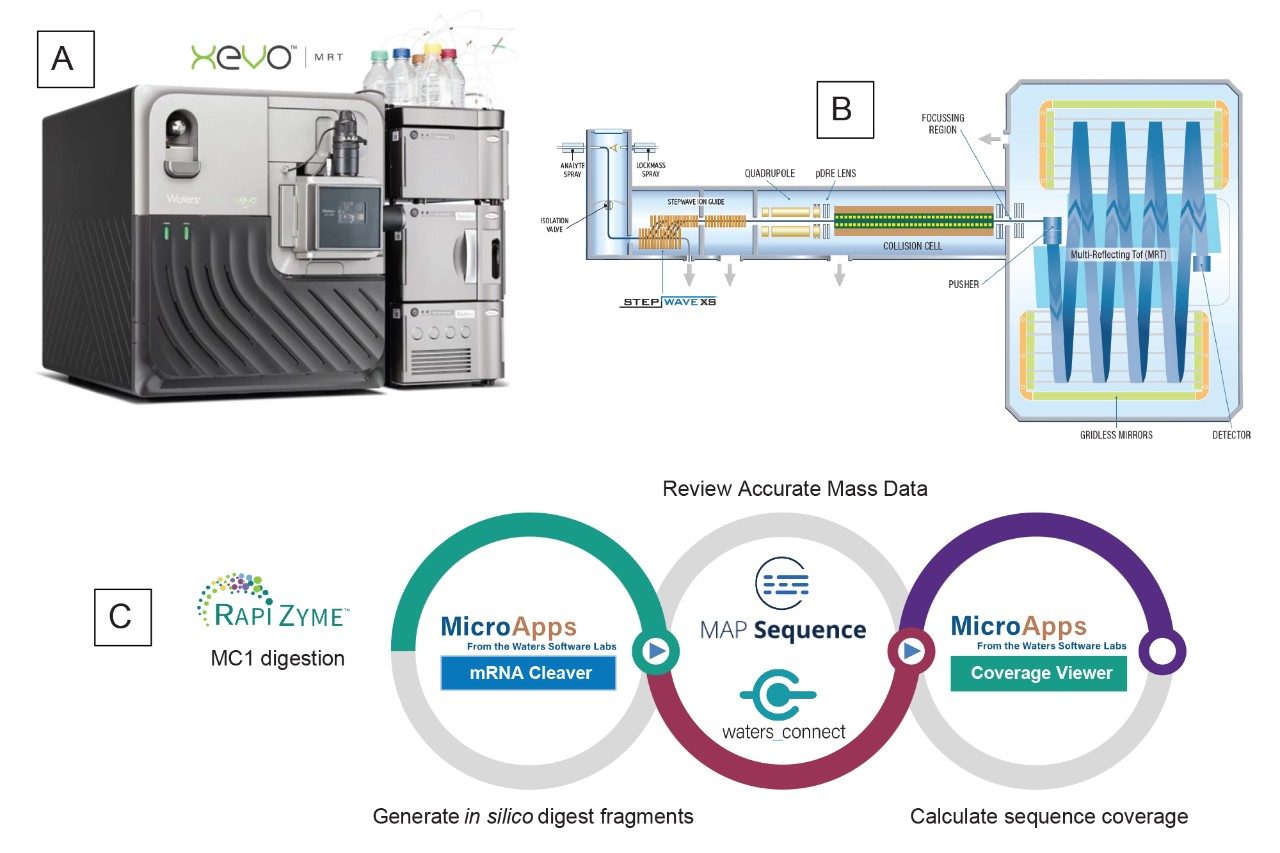
An UPLC-MSE oligo map dataset was acquired for the Waters sgRNA LC-MS oligo standard digested with RapiZyme MC1. The UV and TIC chromatograms (Figure 2) indicate that the MC1 digested products were detected with comparable elution patterns by both UV and MS detection. The UPLC-MSE data was acquired on the Xevo MRT Mass Spectrometer, which uses the multi-reflecting time-of-flight technology (MRT) to achieve exceptional MS resolution, speed, sensitivity and sub-ppm mass accuracy.
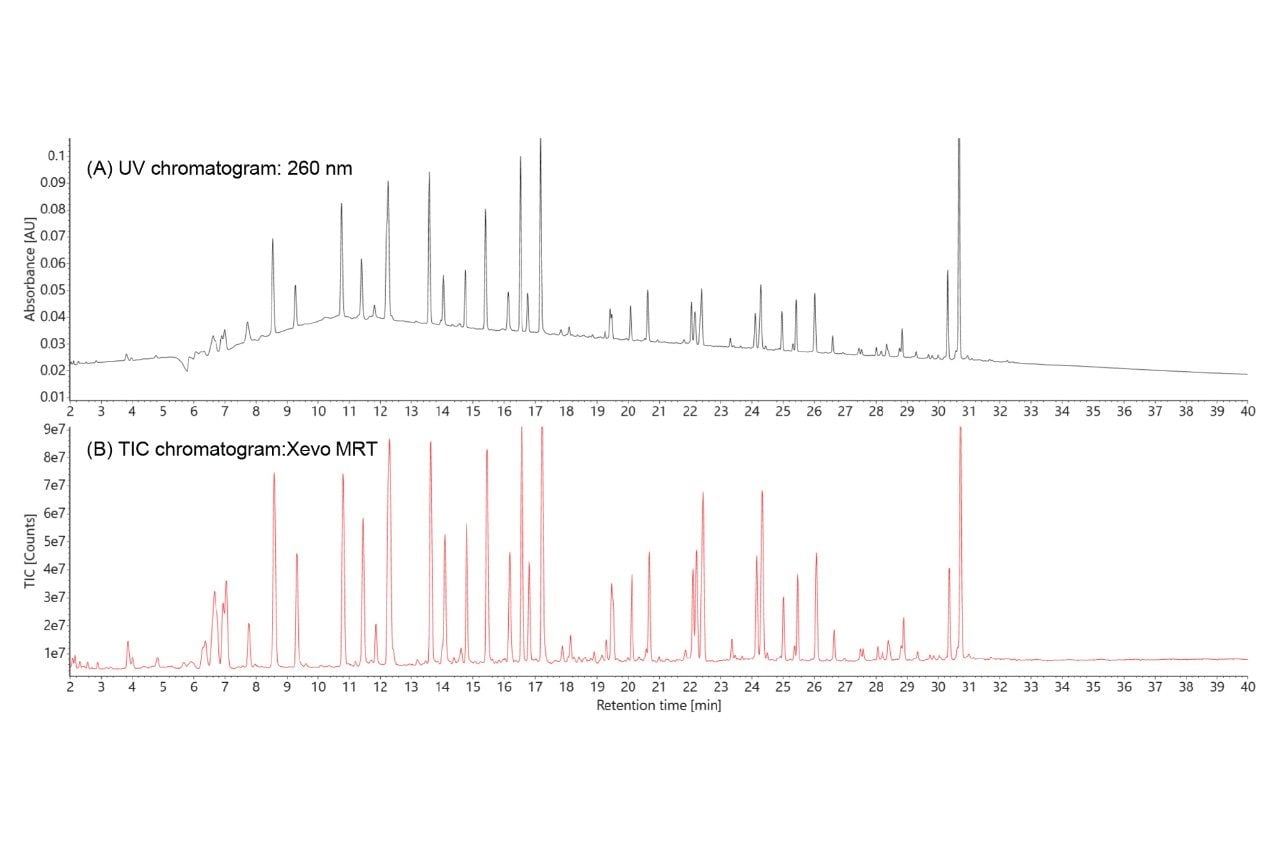
The waters_connect MAP Sequence App (ver 1.0) was used for automated processing of the UPLC-MSE dataset and a portion of the results table is displayed in Figure 3. The green circles displayed next to each digestion product indicate unique assignments based on MS1 (precursor mass) data. Digestion products were assigned with high confidence based on matching the experimentally measured monoisotopic oligonucleotide precursors against the in-silico predicted monoisotopic masses. The mass accuracy observed for all digestion products displayed is in the sub-ppm range (< 1 ppm).
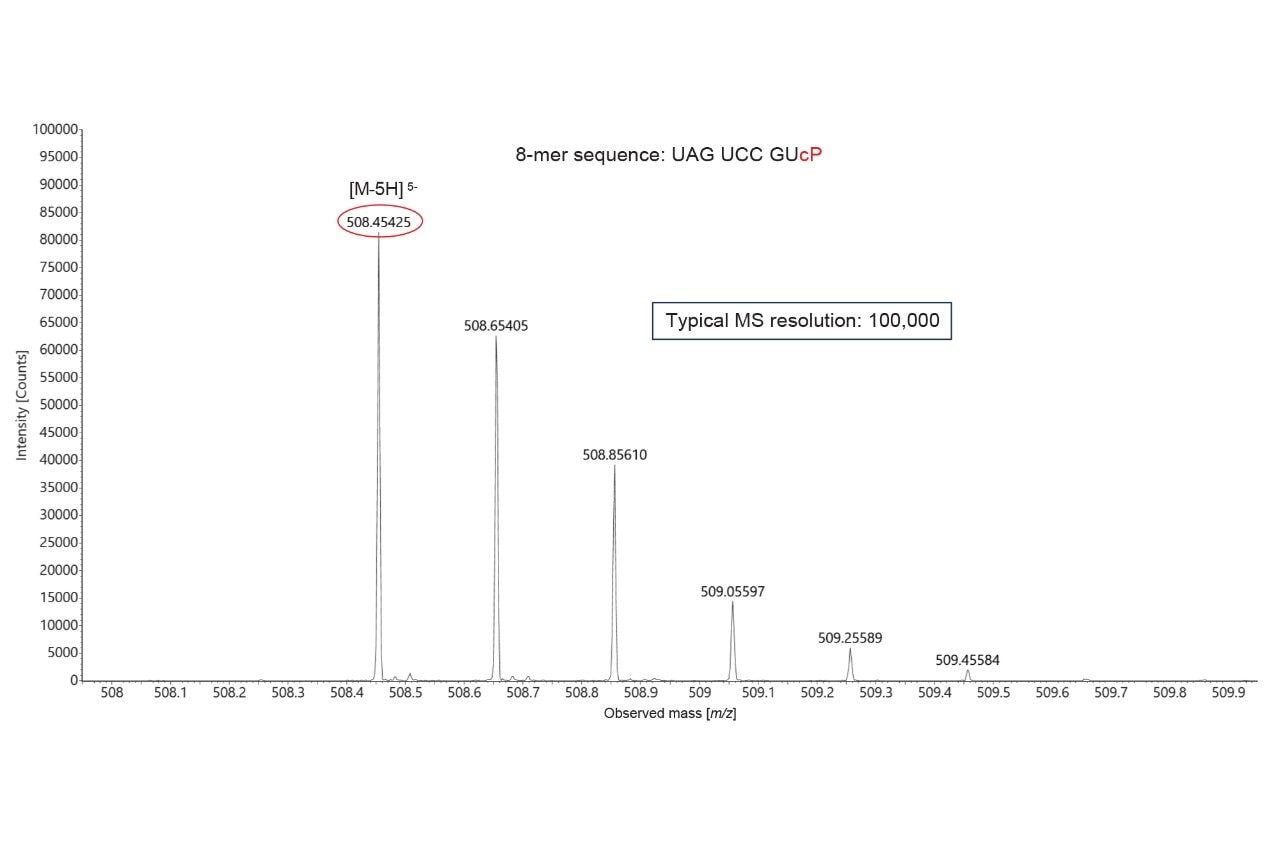
Five different negative charge states (-1 through -5) were detected for the ions corresponding to the highlighted MC1 digestion product (U56:U63, shown in blue), as indicated by the panel labeled “Details” (Figure 3, Right Side). The detailed isotopic distribution of the [M-5H]5- charge state of the U56:U63 (8-mer) MC1 digestion product (Figure 4) displays the MS resolution achieved by the Xevo MRT Mass Spectrometer instrument (~100,000), which fully resolves the multiple isotopes of this (-5) negatively charged precursor.
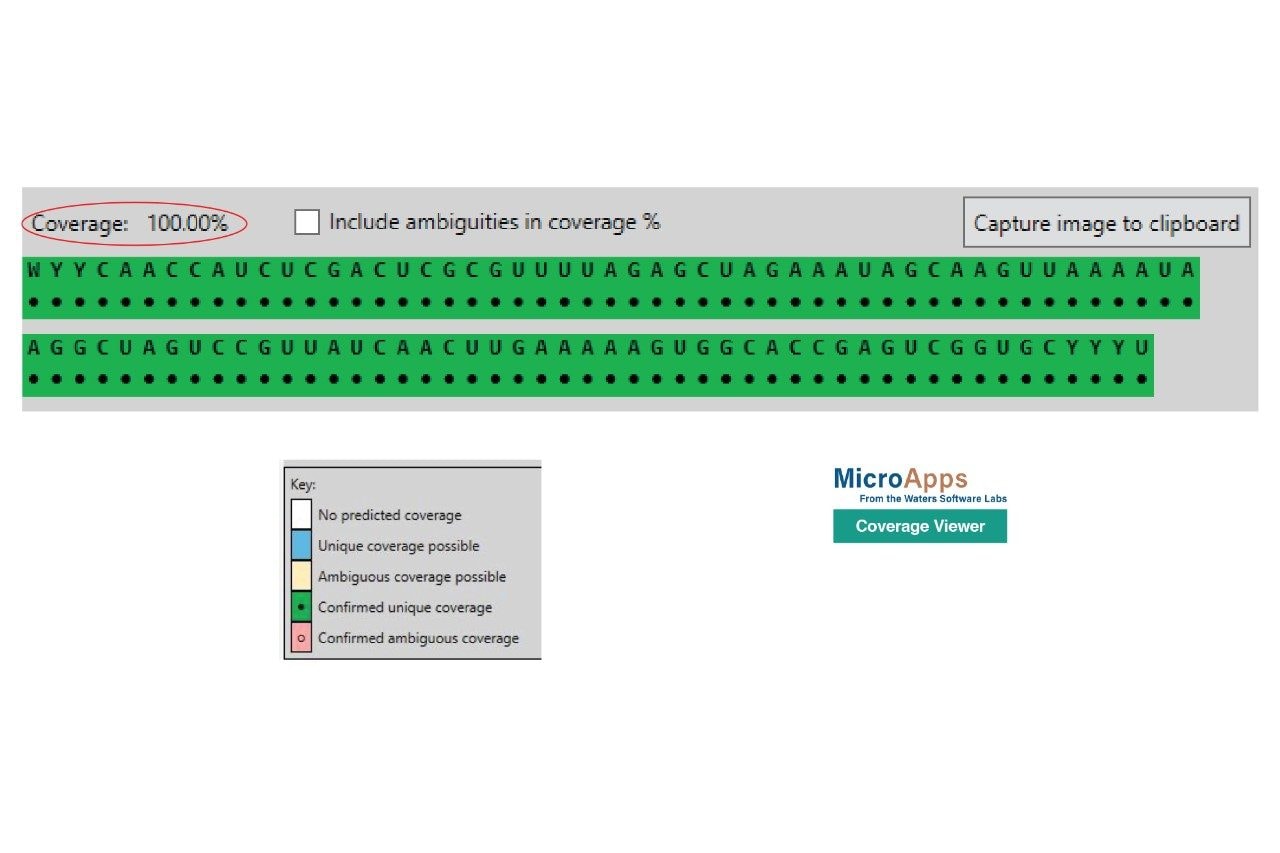
The digested oligo mapping results produced by the Coverage Viewer MicroApp are shown in Figure 5. The Rapizyme MC1 enzyme achieved full (100%) coverage for the Waters sgRNA standard though unique digestion specificity (five distinct dinucleotide cleavage sites), and through its ability to generate controlled missed cleavages. These longer digestion products, generated through 2–4 missed cleavages, are more likely to have unique masses. RapiZyme MC1 cleaves at the 5’-end of uridine residues, with three major cleavage sites (A_U / C_U / U_U) and two minor cleavage sites (C_A / C_G)6-11. MC1 produces mainly digested products ending with a 3’ cyclic phosphate (cP). As a result of these enzymatic features, MC1 exbibits higher coverage, compared to traditional RNase T1 digestion.6–11
Elucidating Ambiguous Sequence Assignments with CONFIRM Sequence
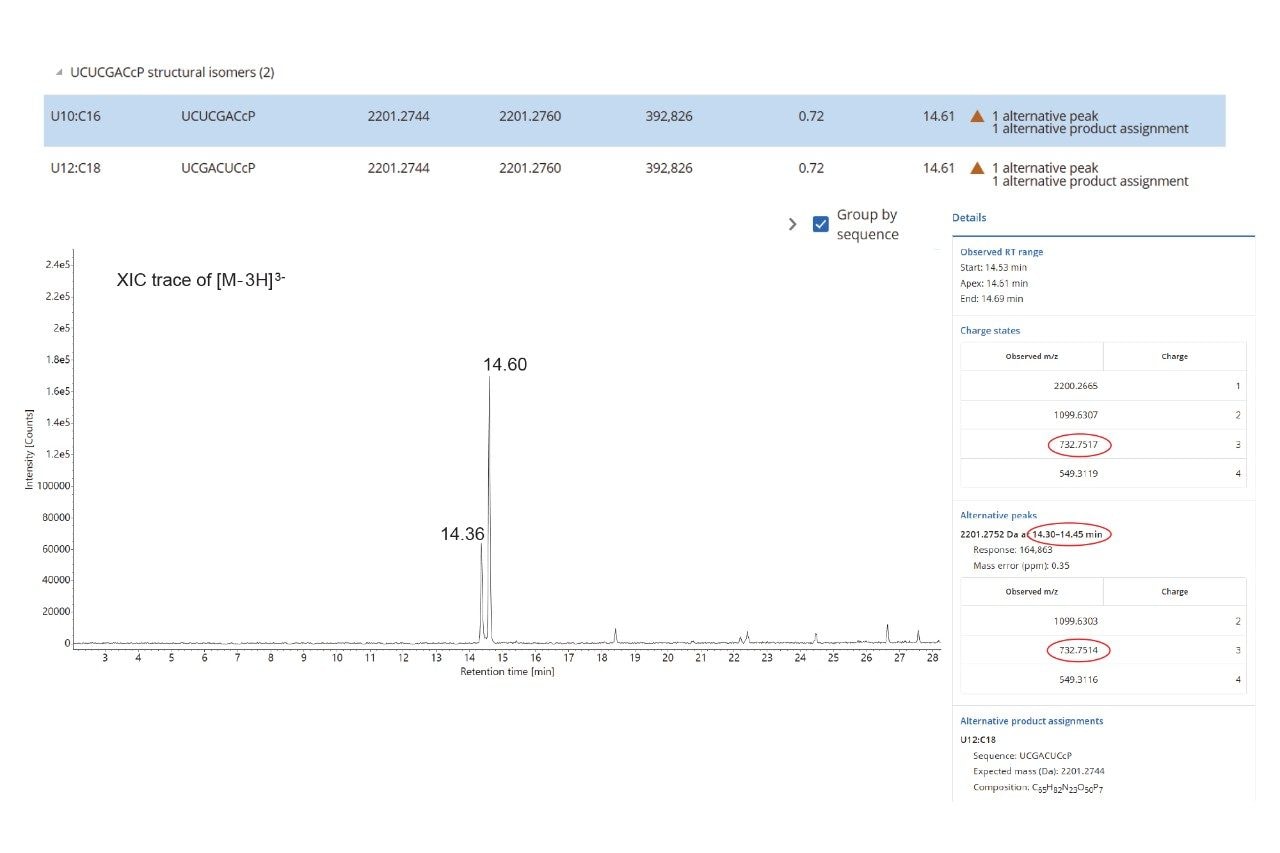
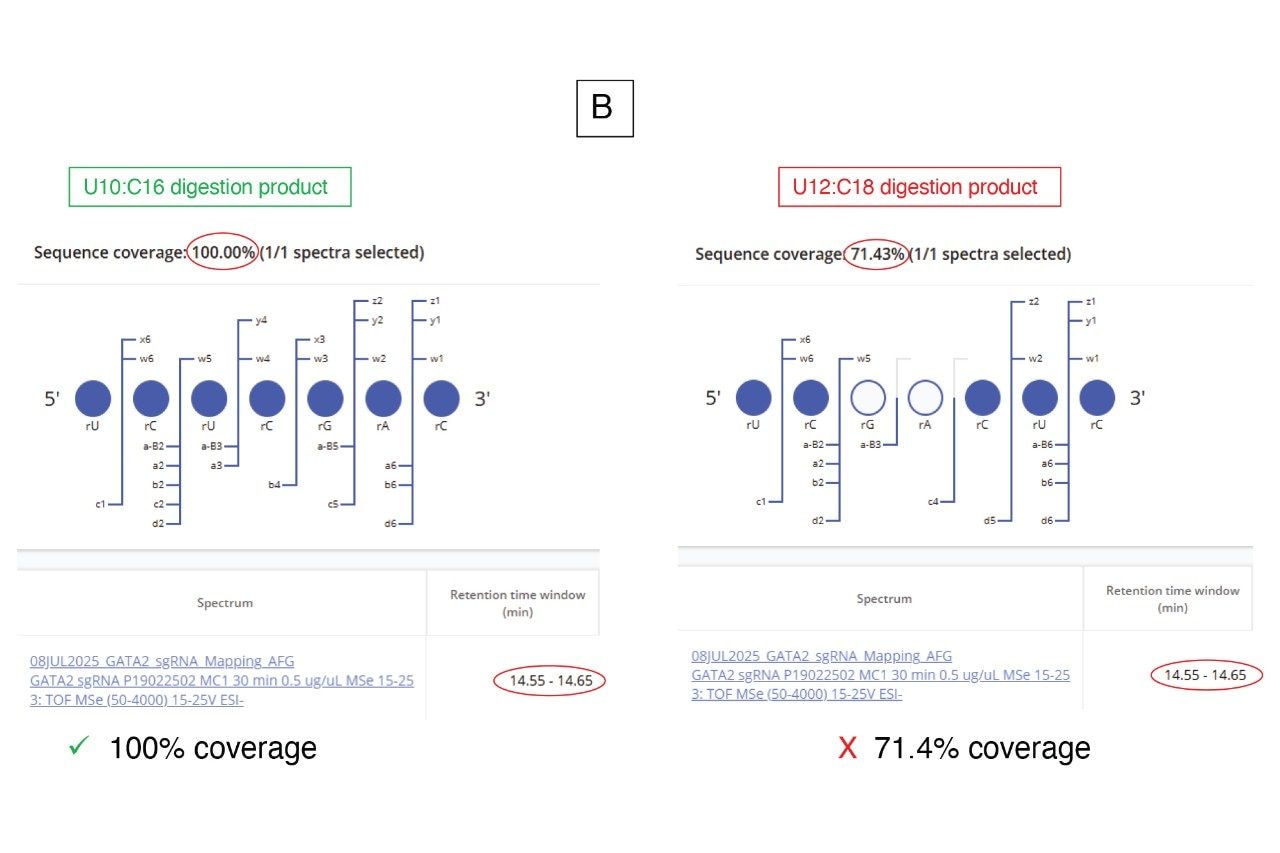
In cases where oligonucleotide sequence assignments are ambiguous, the CONFIRM Sequence waters_connect App12 can be employed to analyze the elevated energy MSE data and assign fragment ions to predict digest product sequences. To illustrate this approach, two 7-mer digested products generated by RapiZyme MC1 treatment of the Waters sgRNA standard are highlighted. These products, U10:C16 (sequence UCU CGA C) and U12:C18 (sequence: UCG ACU C), are structural isomers with identical nucleotide compositions and predicted MS1 mass values.
The MAP Sequence App automatically assigned both sequences (Figure 6, top) by evaluating multiple precursor ions across charge states 1 to 4 and flagged them as “alternative product assignments”. The extracted ion chromatogram (EIC) for the triply charged precursor (m/z = 732.75), also presented in Figure 6, reveals a chromatographic peak doublet with retention times of 14.36 and 14.60 minutes, likely corresponding to these sequence isomers. The MAP Sequence processing result labeled these assignments as having “1 alternative peak”, reporting the most abundant peak at 14.61 minutes as the primary observed retention time.
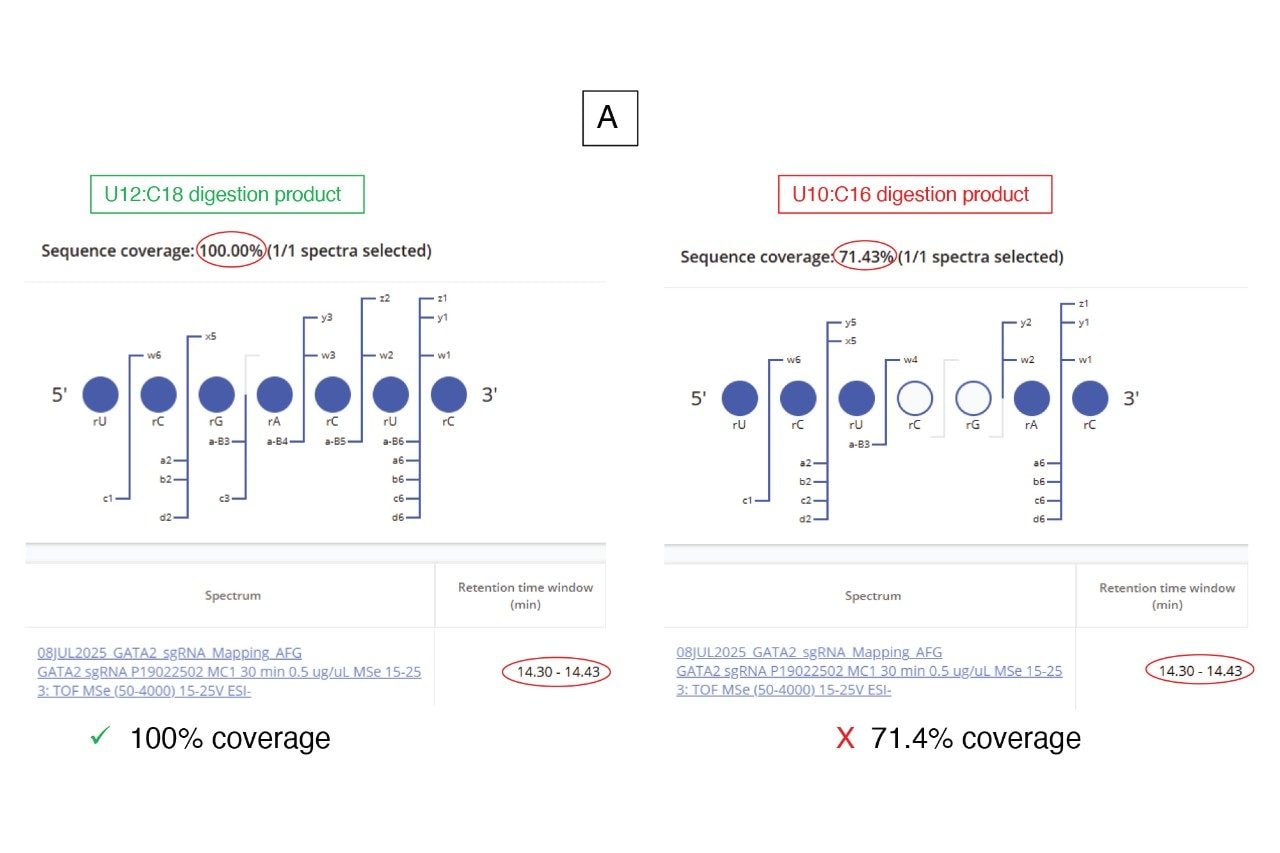
Subsequently, the CONFIRM Sequence App was utilized to automatically assign the fragment ions observed in the elevated energy ESI-MSE spectral channel corresponding to the doublet. Based on this analysis, the peak at 14.36 minutes was clearly assigned to the U12:C18 product, while the peak at 14.60 minutes corresponded to the U10:C16 isomer (Figure 7). In both cases, the elevated energy MSE fragmentation achieved 100% MS2 sequence coverage, providing complete linear sequence verification for each digestion product.
Conclusion
- An informatics workflow, incorporating the waters_connect MAP Sequence App (ver 1.0) was successfully demonstrated for UPLC-MSE digested oligo mapping analysis of sgRNA digests using the Xevo MRT QTOF Mass Spectrometer.
- Confident peak assessments for analysis of sgRNA digests were achieved with the Xevo MRT QTOF Mass Spectrometer, which delivered excellent sensitivity, high resolution MS (100,000), and mass accuracy of less than 1 ppm.
- Complete sgRNA sequence coverage (100%) was achieved by employing RapiZyme MC1, a novel endonuclease introduced by Waters, which provided unique cleavage specificity and predictable missed cleavages to generate longer, unique, overlapping digestion products.
- Oligonucleotide isomeric sequences resulting from enzymatic digestion were effectively differentiated using the waters_connect CONFIRM Sequence App, which matched elevated energy fragment ions patterns to their respective sequences, allowing unambiguous isomer identification.
References
- Jinek M, Chylinsky K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity, Science, 2012, 337, 816–821.
- Jiang F, Doudna JA. CRISPR-Cas9 Structures and Mechanisms, Annu Rev Biophys, 2017, 46, 505–529.
- Ganbaatar U, Liu C. CRISPR-Based COVID-19 Testing: Toward Next Generation Point of Care Diagnostics, Front Cell Infect Microbiol, 2021, 11, https://doi.org/10.3389/fcimb.2021.663949
- LC-MS Analysis of siRNA, Single Guide RNA and Impurities using the BioAccord System with ACQUITY Premier System and New Automated INTACT Mass Application, 2022, Waters application note. 720007546.
- Goyon A, Scott B, Kurita K, Crittenden CM, Shaw D, Lin A, Yehl P, Zhang K. Full Sequencing of CRISPR/Cas9 Single Guide RNA (sgRNA) via Parallel Ribonuclease Digestions and Hydrophilic Interaction Liquid Chromatography High-Resolution Mass Spectrometry Analysis, Anal Chem, 2022, 93, 14792–14801, doi: 10.1021 /acs.analchem. 1c03533.
- RNA Digestion Product Mapping Using an Integrated UPLC-MS and Informatics Workflow, 2024, Waters application note. 720008553.
- Tunable Digestion of RNA Using RapiZyme RNases to Confirm Sequence and Map Modifications, 2024, Waters application note. 720008539.
- Oligo Mapping of mRNA Digests Using a Novel Informatics Workflow, 2025, Waters application note. 720008677.
- Analysis of mRNA Cap Impurity Profiles and Capping Efficiency Using RapiuZyme MC1 Ribonuclease, 2025, Waters application note. 720008793.
- Grunberg S, Wolf EJ, Jin J, Ganatra MD, Becker K, Ruse C, Taron CH, Correa IR, Yigit E. Enhanced Expression and Purification of Nucleotide-specific Ribonucleases MC1 and Cusativin, Protein Expr Purif Acid Res, 2022, 190, 105987, doi: 10.1016/j. pep.2021.105987.
- Thakur P, Atway J, Limbach PA, Addepalli B. RNA Cleavage Properties of Nucleobase-Specific RNase MC1 and Cusativin Are Determined by the Dinucleotide-Binding Interactions in the Enzyme-Active Site, Int J Mol Sci, 2022, 23, 7021.
- CONFIRM Sequence: A waters_connect Application for Sequencing of Synthetic Oligonucleotide and Their Impurities, 2022, Waters application note. 720007677.
720009009, September 2025