For research use only. Not for use in diagnostic procedures.
For research use only. Not for use in diagnostic procedures.
Michael McCullagh1, Stuart Adams1, Jeff Goshawk1, Russell J Mortishire-Smith1, Andrew Tudor1, David Megson 2, Kwadwo Ansong Asante3, Pennante Bruce-Vanderpuije3
1 Waters Corporation, United States
2 Manchester Metropolitan University, UK
3 CSIR-Water Research Institute, Ghana
Published on May 5, 2025
For research use only. Not for use in diagnostic procedures.
Using the SELECT SERIES™ Cyclic™ Ion Mobility System (IMS), data independent analysis (DIA) of human serum of electronic waste (E-waste) handlers has been performed to identify known PFAS and facilitated confirmation of PFAS environmental exposure. In this work we demonstrate the advantages of using the waters_connect™ Pattern Analysis Application data processing workflow. We identify features that are characteristic of PFAS, facilitating visualisation of suspected PFAS that are not incorporated in the PFAS library to enable “known” and “unknown” PFAS putative identification.
Stringent guidelines and legislation have been introduced because of the toxicity associated with PFAS exposure.1–5 The use of DIA to detect and identify PFAS compounds within the environment or to monitor environmental exposure, is driven by the lack of commercially available standards, which are conventionally used to perform targeted analysis. Adverse health conditions resulting from environmental exposure to PFAS have previously been described.6,7 Studies indicate that new alternative PFAS have similar toxicity to banned PFAS, further emphasising the importance of understanding the impact of environmental exposure to both known and unknown PFAS.8
Driven by regulation and a demand for alternative PFAS, the synthesis of new PFAS compounds continues and consequently leads to the presence of not yet known PFAS in the environment together with associated by-products and degradation products. The ubiquitous use of PFAS includes the electronics industry, to improve product quality and performance. The recovery of embedded recyclable electronic components is arduous.9,10 At recycling centres PFAS exposure occurs through inhalation, ingestion, and dermal routes. Atmospheric transport of particulates extends exposure risk beyond recycling centres to surrounding urban areas.
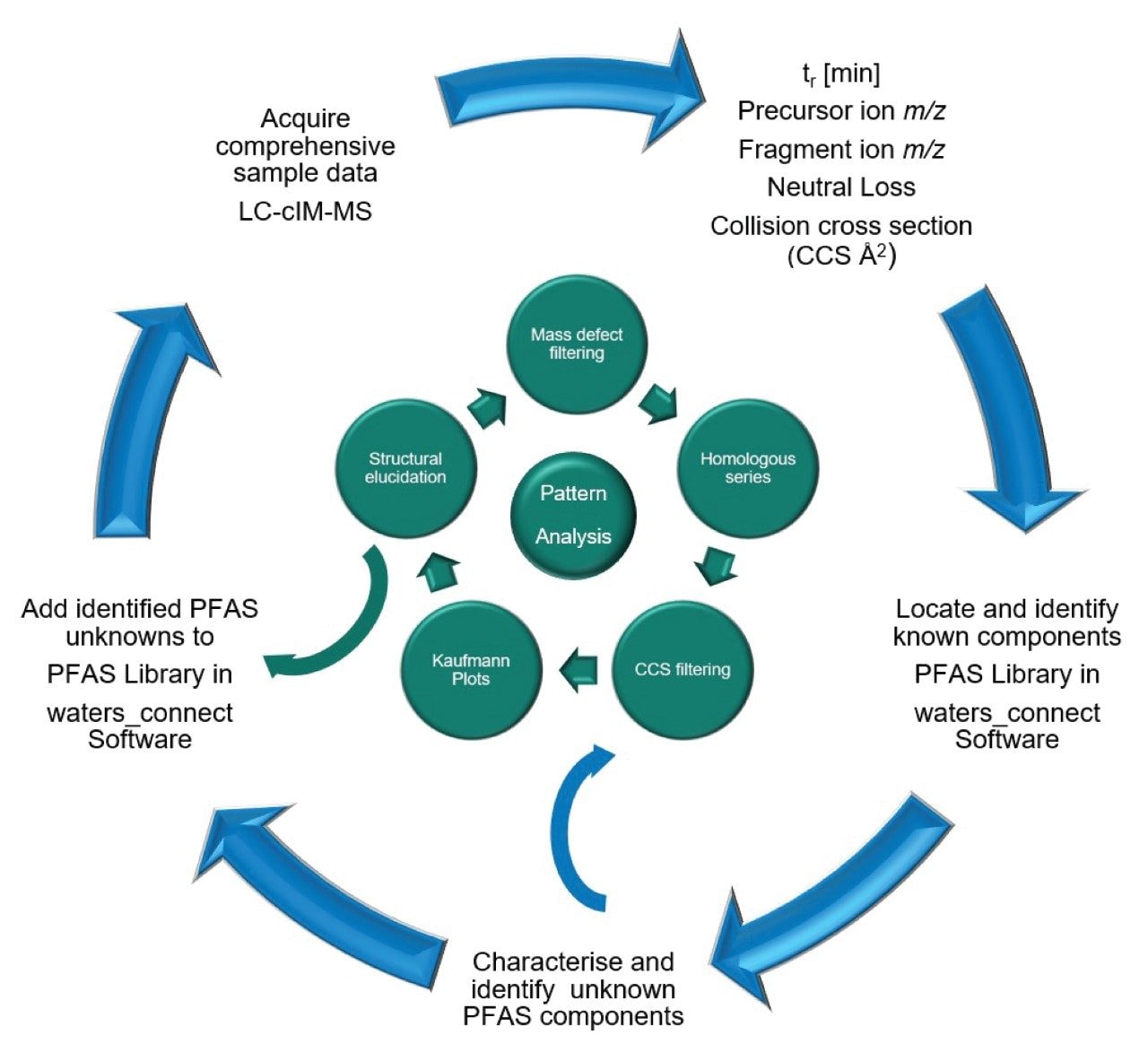
The advantage of a data independent UHPLC-cIM-MS strategy enables identification of known, unknown, emerging PFAS, PFAS precursors, and transformation products.11–13 The highly specific data generated for all detected analytes are analysed using holistic pattern analysis. Using the waters_connect Pattern Analysis Application, PFAS feature patterns are distinguished using information dependent data processing parameters, these are correlated to chemical properties of analytes possessing fluorine and carbon makeup.
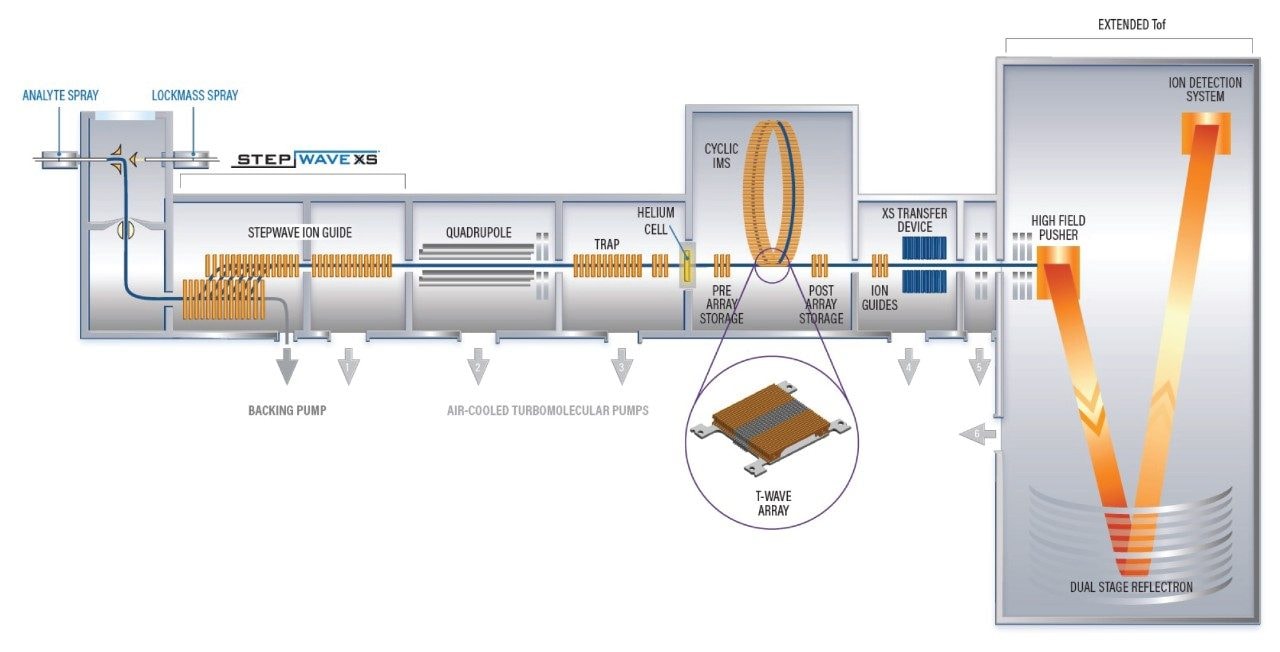
This application note describes the benefits of using the SELECT SERIES Cyclic IMS (see Figure 1) and the waters_connect Pattern Analysis Application to investigate a subset of data acquired from the analysis of the human serum taken from Ghanaian E-waste handlers who may have been exposed to environmental PFAS.
E-waste handler anonymised human serum samples.
Anonymised human serum samples were extracted using SPE 96-well µElution plates, containing a polymeric reversed-phase, weak anion exchange mixed-mode sorbent.14
|
LC system: |
ACQUITY™ Premier System modified with PFAS Kit and Atlantis™ Premier BEH™ C18 AX Isolator Column, 2.1 x 50 mm, 5 µm (p/n: 186010926).15 |
|
Column: |
ACQUITY UPLC™ BEH C18 Column (100 mm x 2.1 mm, 1.8 µm) |
|
Column temperature: |
35 °C |
|
Sample temperature: |
6 °C |
|
Injection volume: |
5 µL |
|
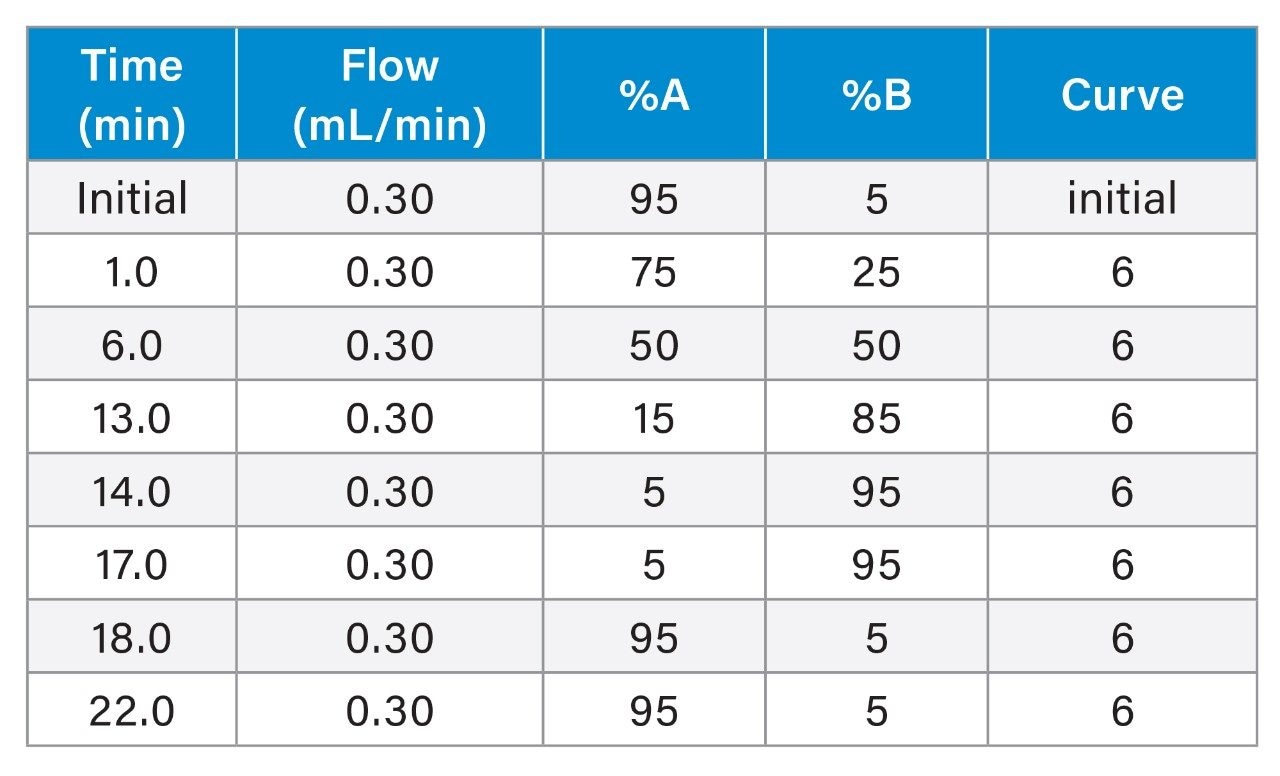
Flow rate: |
0.3 mL/min |
|
Mobile phase A: |
95 H2O (2 mM ammonium acetate):5 MeOH |
|
Mobile phase B: |
MeOH (2 mM ammonium acetate) |
|
Ionization mode: |
ES- |
|
Acquisition range: |
m/z 50–1200 (HDMSE) |
|
Capillary voltage: |
0.5 kV |
|
Collision energy: |
20–45 eV |
|
Cone voltage: |
10 V |
|
Desolvation temperature: |
350 °C |
|
Source temperature: |
100 °C |
|
Acquisition rate: |
0.16 seconds (HDMSE) R~65 |
|
Chromatography and MS: |
MassLynx™ 4.2 SCN1026 Software |
|
Informatics: |
waters_connect 3.1.0.243 Software waters_connect Pattern Analysis Application 1.0 |
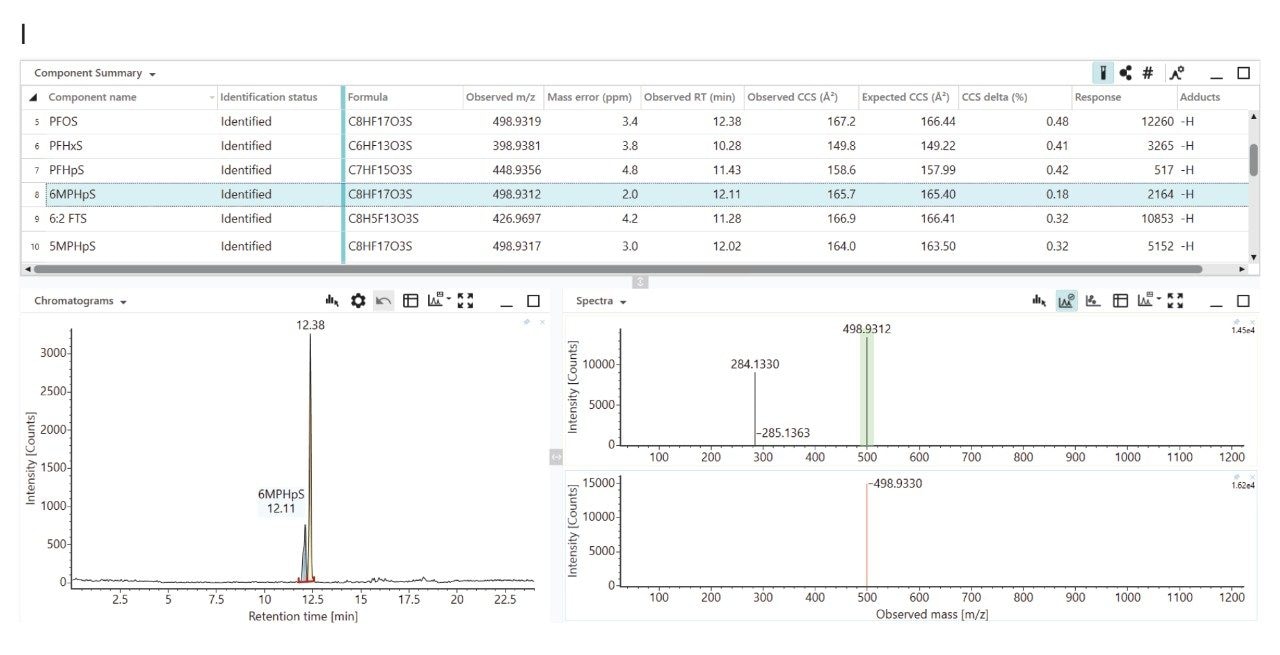
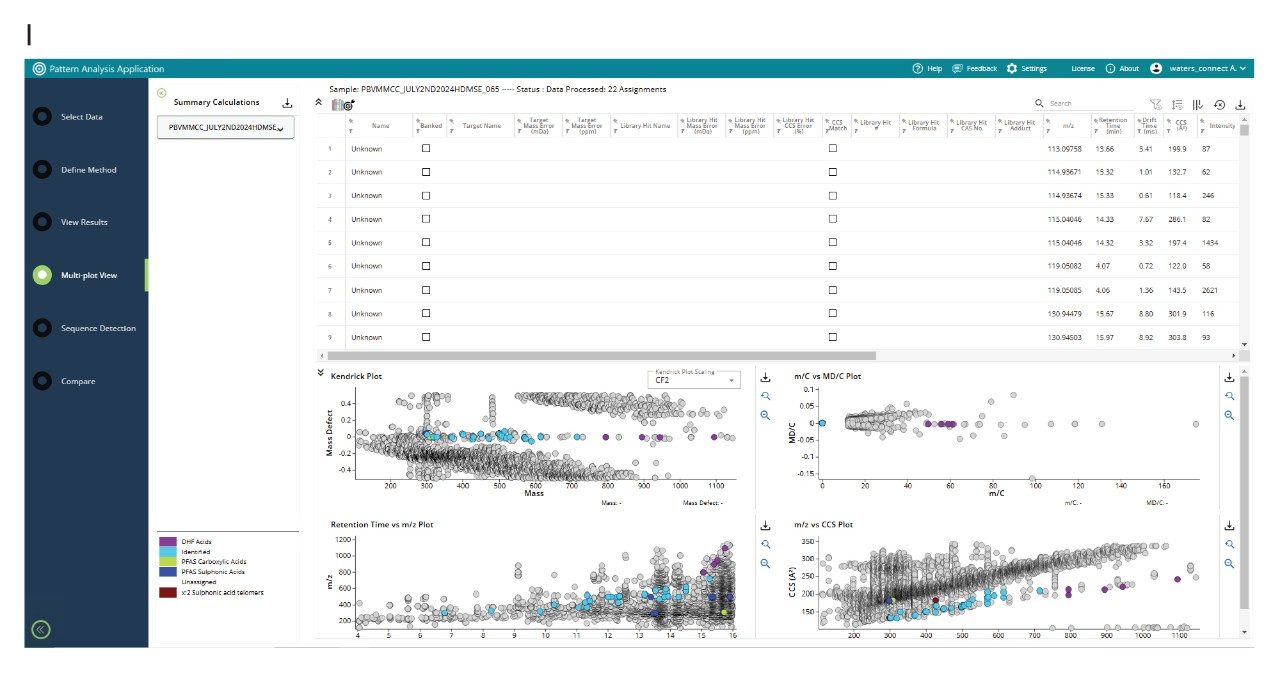
Non-targeted UHPLC-cIM-MS was used to analyse anonymised human serum samples of Ghanaian E-waste handlers and confirmed environmental exposure to PFAS, identifying PFAS including 6:2 FTS, PFHxS, PFHpS, and PFOS isomers (L-PFOS, 5MPHpS, 6MPHpS) with Δ CCS <0.5% compared to the PFAS library values (see Figure 2 (I)). The results showed a Δ CCS of less than 0.5% when compared to PFAS library values, demonstrating how data independent analysis and post-acquisition information dependent data processing can be used to confidently determine environmental exposure to known PFAS.
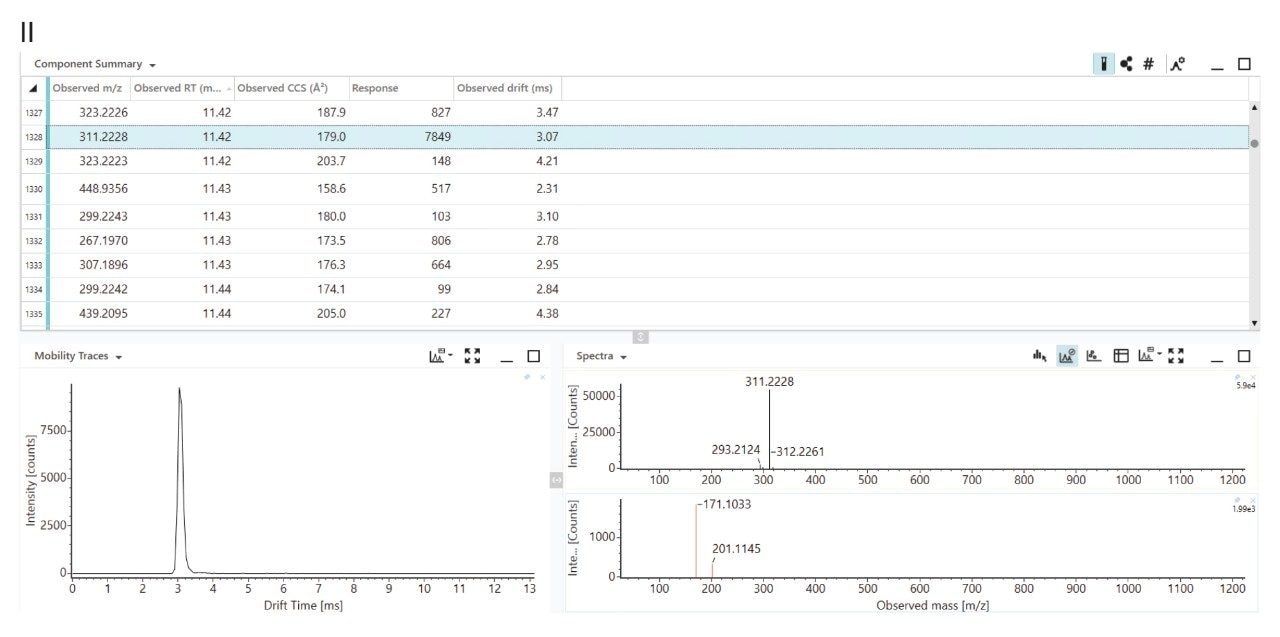
Although developed countries have implemented stringent guidelines for PFAS, developing countries continue production and often lack regulations and mechanisms to address emerging PFAS. The United States Environmental Protection Agency (USEPA) maintains a database of approximately 14,800 PFAS analytes.16 DIA affords opportunity to use a holistic approach to determine exposure to novel and emerging PFAS, in complex matrices such as human serum, soil and food, which can contain 1000’s of small molecule analytes within the mass distribution range of PFAS. When performing DIA, precursor ion m/z, tr, and CCS values are acquired for all analytes detected (Figure 2 (II)). Using the acquisition and data processing workflow shown in Figure 3, holistic pattern analysis can be used to characterise information and detect “known unknowns” and “unknown” PFAS within the data, including PFAS for which analytical standards are not available and those that are not present in the PFAS library.
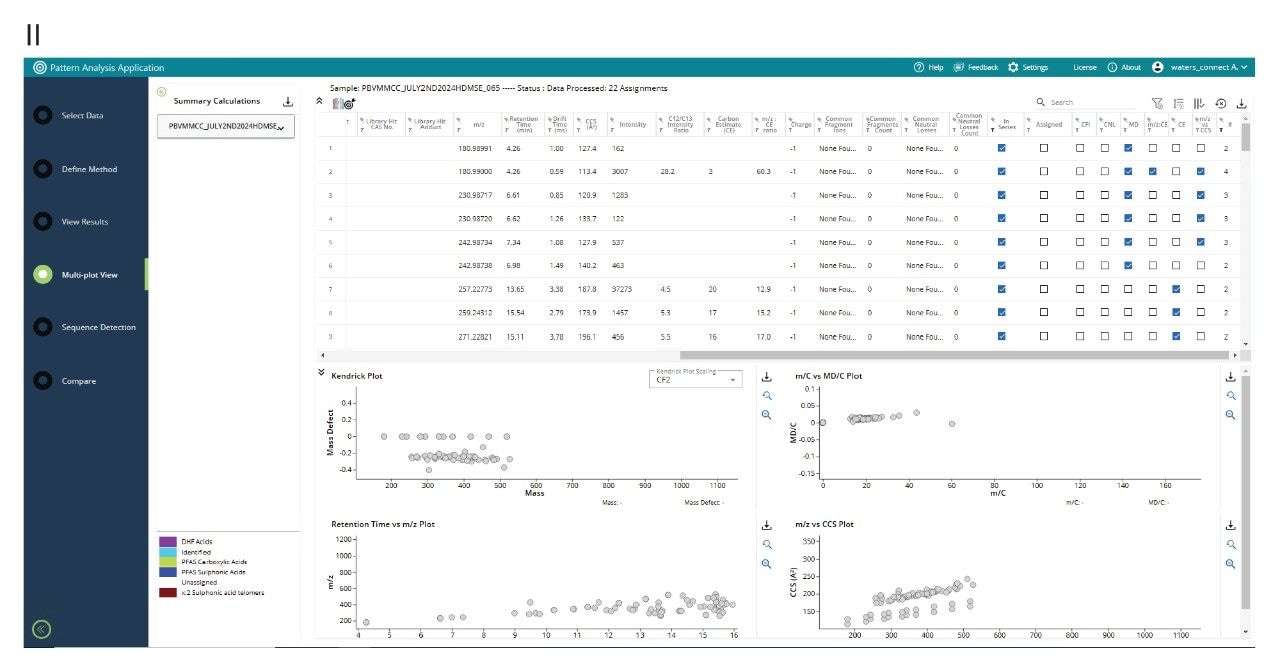
The Pattern Analysis Application derives additional downstream outcomes from the primary data, where criteria are set to perform additional data processing of the “unknowns” data imported from waters_connect Software. Kaufmann proposed “Kaufmann plots” as a single step non-targeted strategy to detect PFAS using a plot of (m/C versus md/C), where mass (m), carbon number (C), mass defect (md) with a two-dimensional graph (m/C versus md/C), to visualise suspected PFAS.17 Kendrick mass defect is increasingly used in environmental sciences to differentiate natural organic matter, from groups of environmental contaminants and can facilitate selection of potential features to be retained for PFAS suspect screening.18,19 Here, we normalise the Kendrick mass defect to the CF2 repeat unit. As a result, any features differing by this mass form a straight line, aiding the differentiation between natural compounds and anthropogenic PFAS compounds. Additionally, distinctive PFAS mass/CCS trendlines can be used to identify suspected PFAS features.20 Using the waters_connect Pattern Analysis Application, holistic PFAS pattern analysis has been performed using a defined CF2 repeat unit, designated PFAS repeat unit end groups, component spectrum, Kendrick mass defect, Kaufmann plots (m/C vs md/C), retention time vs m/z plot and m/z vs CCS plot (see Figure 4 (I)). Additional filtering parameters, such as common fragment ion, common neutral loss, CCS, and (CF2)n homologous series were applied. The post-processing and filtering results are presented in Figure 4 (II). The results for a match in the “waters_connect Pattern Analysis Application PFAS library” and an “unknown” PFAS detected in E-waste handler human serum are shown in Figure 5. The waters_connect Pattern Analysis Application PFAS library has been constructed using content of the CompTox Chemical Dashboard vs2.5.2.16 It contains library hit name, elemental composition and theoretical CCS values generated using machine learning (PFASSTRUCTV5).21
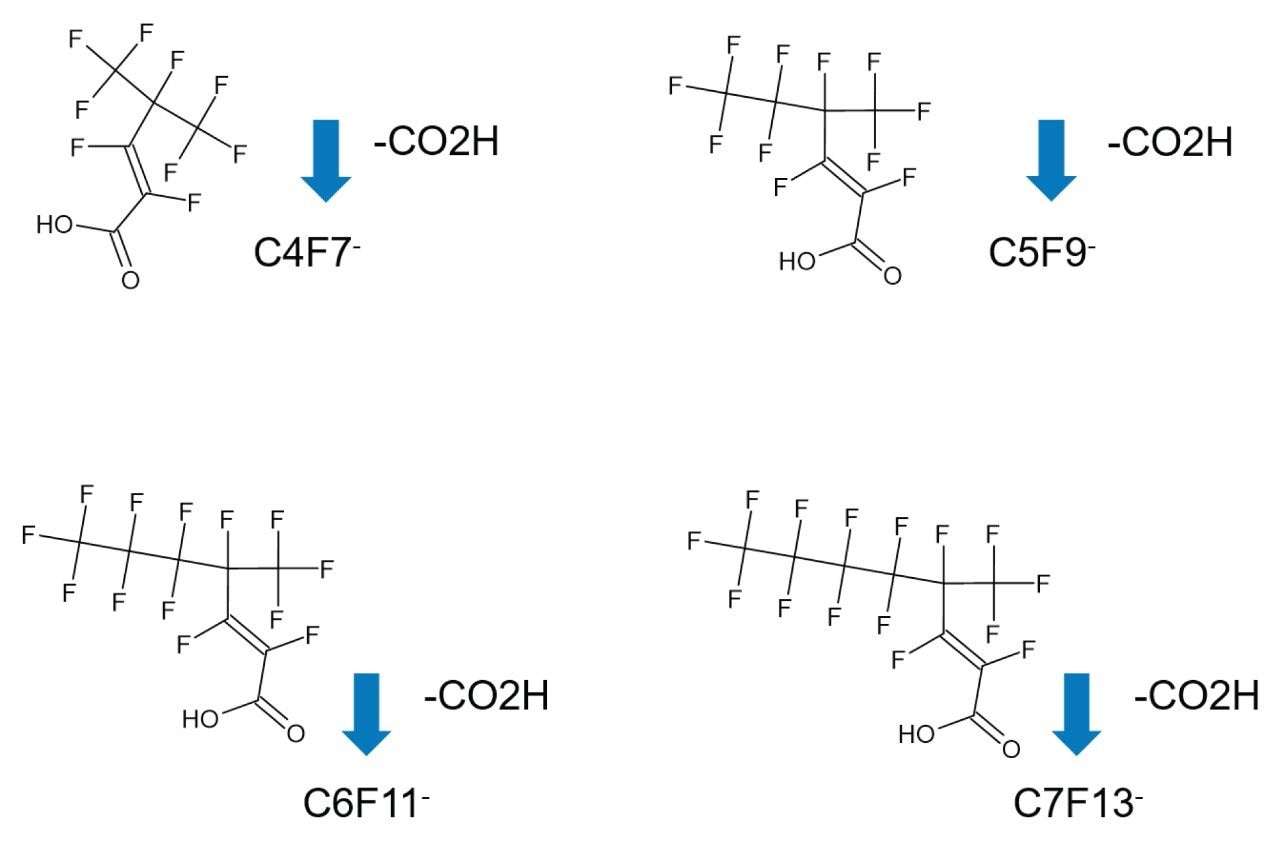
A homologous series of unsaturated PFAS anions have been observed (C4F7-, C5F9-, C6F7-, and C7F13-); they diverge from the waters_connect Software PFAS library matches for detected known PFAS, excluding potential false assignment of fragment ions of labile PFAS. Despite the absence of fragmentation information at low intensity responses, a characteristic CCS fingerprint of a labile PFCA species was obtained. By combining elemental composition, information from the waters_connect Pattern Analysis Application PFAS library, and the observed CCS fingerprint, it is possible to putatively assign the four unknown PFAS species as unsaturated PFCAs, see Figure 6. Barola et al. have previously investigated unknown PFAS and putatively assigned unsaturated sulphonic acid, carboxylic and PFAS structures.22 The mechanistic degradation of PFECAs, including Hexafluoro-4-(trifluoromethyl) pent-2-enoic acid (PFMeUPA) have also been investigated.23,24 PFMeUPA is used as a fluorinated solvent and cleaning agent in the semiconductor industry. Semiconductor electrical properties are foundational for computers, electronic devices, and everyday electrical appliances; it is possible we have identified a perfluoroalkyl homologous series containing perfluoro-(4-methyl-2-pentenoic acid). Although branched unsaturated PFCAs, such as perfluoro-(4,4-dimethylpent-2-enoic acid), may have been observed, the retention time information from the waters_connect Pattern Analysis Application indicates the observation of an unsaturated PFCA homologous series. Additional confirmation may be demonstrated using concentrated samples or analytical standards.

Known PFAS have been identified in anonymised human serum of E-waste handlers using highly specific non-targeted UHPLC-cIM-MS data, incorporating retention time, CCS values, and accurate mass measurements. Applying the waters_connect Pattern Analysis Application facilitated a holistic approach to discriminate unknown PFAS related analytes from thousands of matrix components. This approach uses the distinctive mass spectral properties of perfluorinated analytes to disclose suspected PFAS with high certainty. It has enabled the putative assignment of a homologous PFAS series not present in the PFAS library. Identification of unknown PFAS further confirms the advantage of non-targeted analysis, where highly specific data is generated for all analytes detected. UHPLC-cIM-MS and holistic pattern analysis using the waters_connect Pattern Analysis Application streamlines data analysis for monitoring known and unknown PFAS resulting from environmental exposure.
Ethical Approval: This study received ethical approval from the Ghana Health Service Ethics Review Committee (ref: GHS-ERC 023/01/25) on February 9th, 2025.
Funding Source: This study was financially supported by the Schlumberger Faculty for the Future for Women from Developing Countries in 2016, the TWAS-UNESCO/SG-NAPI/BMBF grant under the agreement number 4500454086, and the Royal Society of Chemistry (RSC) M19–198.
The funding organizations- Schlumberger, TWAS-UNESCO/SG-NAPI/BMBF, and RSC had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Waters, SELECT SERIES, Cyclic, ACQUITY, UPLC, BEH, waters_connect, Atlantis, and MassLynx are trademarks of Waters Technologies Corporation.
We thank the authors Pennante Bruce-Vanderpuije, Kwadwo Ansong Asante from the CSIR-Water Research Institute, Accra, Ghana, and David Megson for their valuable contributions to this application note.
720008784, April 2025