For research use only. Not for use in diagnostic procedures.
Symphony Software is a client/server application that is triggered by a MassLynx acquisition and allows the automation of one, or several, data handling or processing functions in a sequence. It is built with efficiency, flexibility and creativity in mind and enables laboratories to extract the maximum value from LC-MS instrumentation.
By using Symphony Data Pipline Software, a considerable 42% time savings in the overall experiment time was illustrated.
By using the new Symphony Data Pipeline Software, we illustrate a considerable 42% time savings in the overall experimental time in the DIA LC-MS acquisition and data analysis of five complex samples examined for proteome content.
Symphony Data Pipeline (Figure 1) is a newly introduced software product which enables the automation of data processing functions in a sequence following a MassLynx acquisition. There is great flexibility in the types of tasks or functions that can be used. The general guideline is anything that can be executed from a Windows command line prompt can be utilized. The aim of Symphony is to produce flexible data processing routines, save operator time, and allow the extraction of maximum value from LC-MS instrumentation. In this experiment we illustrate the use of Symphony, a file copy task, and component modules of ProteinLynx GlobalSERVER (PLGS) in the analysis of five complex samples being examined for proteome content.
HeLa tryptic digest were kindly provided by Institute of Chemistry, Academia Sinica.
|
LC system: |
ACQUITY UPLC M-Class |
|
Column(s): |
5 μm Symmetry C18 180 μm x 20 mm 2G trap column and 1.8 μm HSS T3 C18 75 μm x 150 mm NanoEase analytical column |
|
Column temperature: |
35 °C |
|
Flow rate: |
300 nL/min |
|
Mobile phase: |
water (0.1% formic acid) (A) and acetonitrile (0.1% formic acid) (B) |
|
Gradient: |
3% to 40% B in 90 min, re-equilibration time 30 mins, total run time 120 mins |
|
Injection volume: |
1 μL |
|
MS system: |
SYNAPT G2-Si |
|
Ionization mode: |
ESI (+) at 3.2 kV |
|
Cone voltage: |
30 V |
|
Acquisition mode: |
(U)(H)DMSE ion mobility assisted DIA1 |
|
Acquisition range: |
50 m/z to 2000 m/z both functions (low and elevated energy) |
|
Acquisition rate: |
Low and elevated energy functions at 0.5s |
|
Collision energy: |
5 eV (low energy function) and from 19 eV to 45 eV (elevated energy function) |
|
Resolution: |
25,000 FWHM |
|
IMS T-wave velocity: |
700 m/s |
|
IMS T-Wave pulse height: |
40 V |
The LC-MS data was processed with Symphony Data Pipeline v1 which was configured with the following tasks: Data Transfer (a batch file which uses Windows Robocopy to transfer files between two PC locations); Apex3D (the peak detection component of PLGS V3.0.3); Peptide3D (the de-isotoping and charge state deconvolution component of PLGS); IA (Ion Accounting, the database search component of PLGS).
The task sequence can be seen in Figure 2. Processing was performed using a Lenovo D20 PC (4 processors, 24GB RAM, NVIDIA C2050 GPU).

Five replicate injections of HeLa tryptic digest were injected and LC-MS acquisitions were performed as described. The timeline for a conventional protocol without using Symphony for this type of experiment can be seen in Figure 3. MS acquisition occurred during the 90 min LC gradient period but was switched off to save PC disk space during the LC column re-equilibration time. At the end of five acquisitions, ~100 GB of data was manually transferred as a single block from the instrument-linked PC to a processing PC (~45 min) and manually processed through the PLGS workflow. The total time for LC-MS acquisition and processing for all five samples was 19 hrs 30 min.
An alternative experimental strategy using Symphony is also illustrated in Figure 3. LC-MS data was automatically transferred from the instrument-linked PC to the processing PC during the LC re-equilibration phase and data processing was automatically performed. At the end of the five acquisitions, all data had already been processed. Total time was now only 11 hrs 20 min.
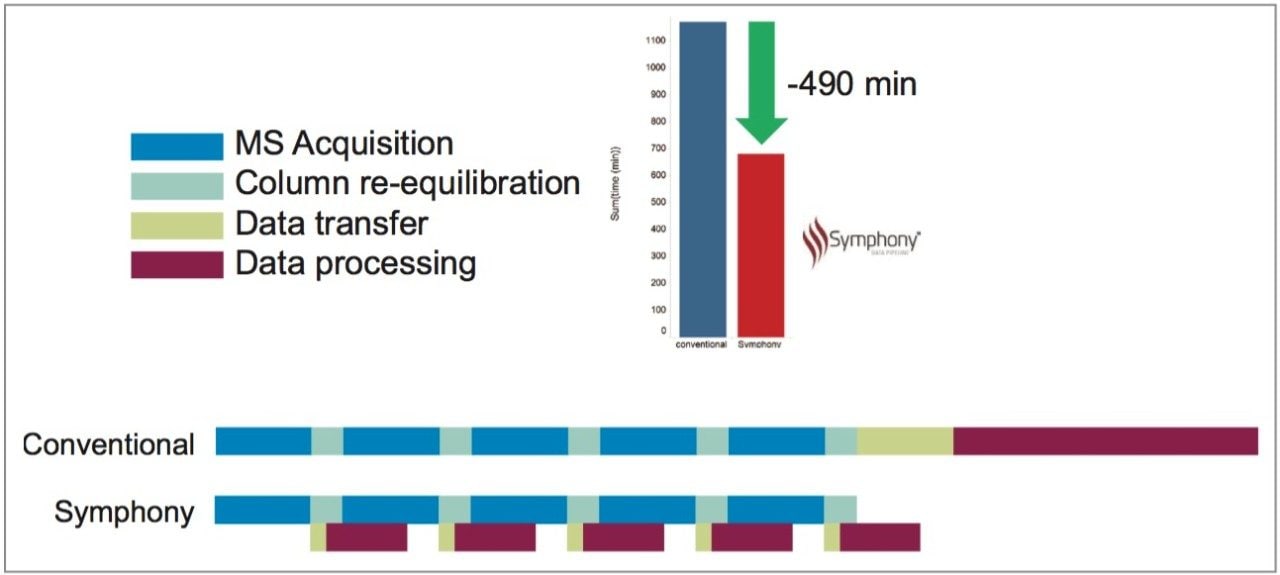
We demonstrate here the use of a new software tool called Symphony Data Pipeline for the streamlining of data processing from a proteomics experiment. LC-MS data was automatically transferred as soon as it was available from the instrument-linked to a processing PC and subsequently processed through a series of PLGS tasks. In using this Symphony workflow on a relatively small set of five samples, we have demonstrated a time savings of 490 mins, 8.2 hrs, 42%, or an entire working day from an experiment normally taking ~2.5 days.
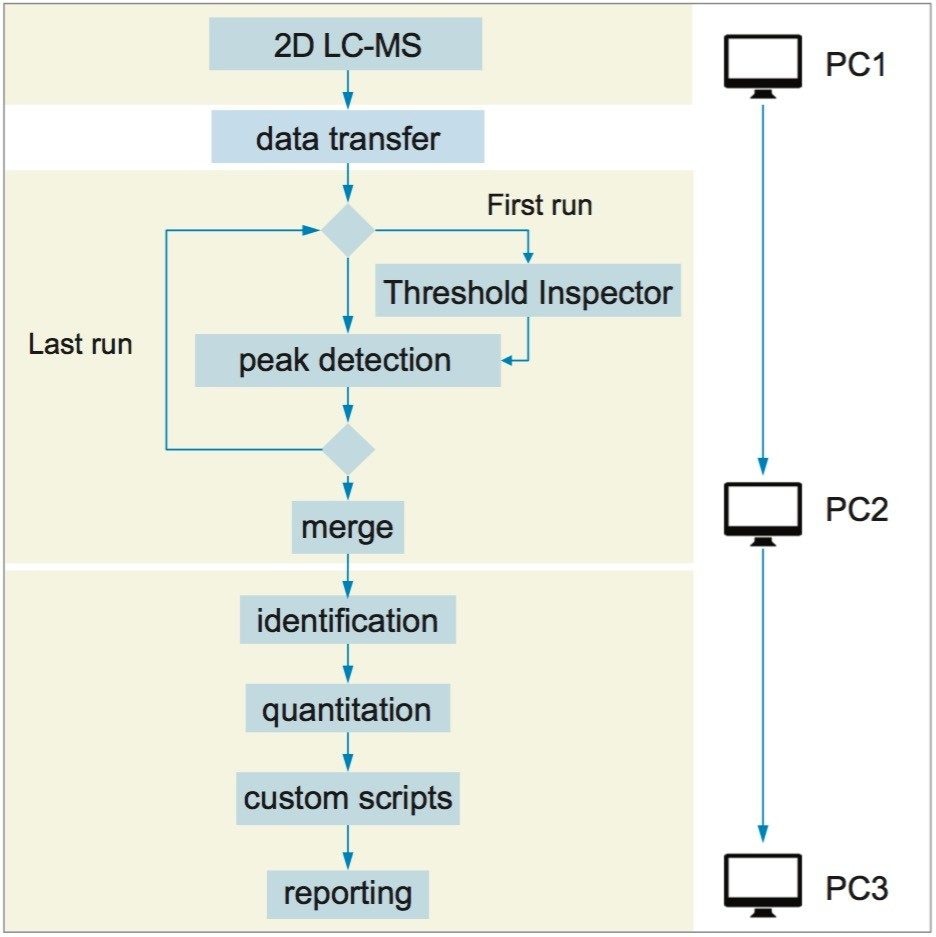
The Symphony pipeline illustrated above is a very simple one with only four tasks. Symphony can also easily accommodate much more advanced configurations such as that shown in Figure 4. Here data from a 2D-LC experiment was processed using a Threshold Inspector task for optimized peak detection parameters, along with a Merge task to pool data from multiple LC fractions. Custom scripting and reporting was accommodated and the whole pipeline was run across three separate PCs.
Symphony Data Pipeline Software provides a flexible platform for the processing of LC-MS data and through automation, can lead to significant time savings. Here we illustrate these benefits through the use of a pipeline for the processing of proteomics data from a complex proteome sample. Other applications of Symphony can be found on the Symphony User Community at http://forums.waters.com/groups/symphony.
720005784, August 2016